Honors
- 1994 National Science Foundation Young Investigator Award
- 1994 Packard Fellowship in Science and Engineering
- 1995 Beckman Young Investigator Award
- 2000 – 2005 HHMI Associate Investigator
- 2000 Protein Society Young Investigator Award
- 2002 International Society for Computational Biology Overton Prize
- 2003 – 2004 Director, Biomolecular Structure and Design Graduate Program (BMSD)
- 2004 AAAS Newcomb–Cleveland Prize
- 2004 Foresight Institute Feynman Prize
- 2005 – HHMI Investigator
- 2006 – National Academy of Sciences
- 2008 Sackler Prize in Biophysics
- 2009 – American Academy of Sciences
- 2011 University of Washington Inventor of the Year Award
- 2012 Biochemical Society Centenary Award
- 2014 American Chemical Society David Perlman Memorial Award
- 2017 – Henrietta and Aubrey Davis Endowed Professorship in Biochemistry
- 2018 Solvay Public Lecture
- 2018 Protein Society Hans Neurath Award
- 2019 The Audacious Project Recipient
- 2020 – Fellow, American Institute for Medical and Biological Engineering
- 2021 The Breakthrough Prize, Life Sciences
- 2023 Frontiers of Knowledge Award in Biology and Biomedicine
Baker Group website
Research
Our research is focused on the prediction and design of protein structures, protein folding mechanisms, protein-protein interactions, protein-nucleotide interactions, and protein-ligand interactions. Our approach is to use experiments to understand the fundamental principles underlying these problems, to develop simple computational models based on these insights, and to test the models through structure prediction and design. We strive to continually improve our methodology by iterating between computational and experimental studies.
The successful application of our computational prediction and design method, ROSETTA, is illustrated in a few recent examples:
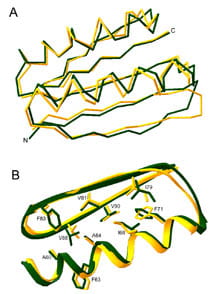
Comparisons of the Top7 design (green) and x-ray structure (yellow). (A) C-alpha overlay. (B) Overlay of core sidechains in the C-terminal portion.
(i) We used computational protein design methods to create an artificial globular protein with a novel fold. Experimental characterization of Top7 showed that it is extremely stable, and the x-ray crystal structure is strikingly close to the design model. These results suggest that new proteins can be designed with atomic level accuracy, and current work is aimed at using these techniques to design new proteins with novel functions.
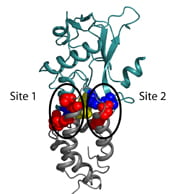
Backbone schematic of the colicin E7 DNase (teal) / Im7 Immunity protein (grey) complex. Important interfacial residues are shown in spacefill (E7 in red, Im7 in blue, conserved Tyr-Tyr motif in yellow).
(ii) We have redesigned protein-protein interaction specificity and demonstrated that the specificity changes hold both in vitro and in vivo.
(iii) Ab initio protein structure prediction. We produced de novo structure predictions of unprecedented accuracy in the recent CASP4 and CASP5 international blind tests of protein structure prediction methods. A more detailed description of our research can be found at https://bakerlab.org/
Publications: