Genetic ‘Hot Spots’ Linked to Developmental Disabilities
by Kris Freeman
Evan Eichler uses a DNA microarray scanner to find large,
spontaneous mutation in chromosomal hot spots and link
them to a range of developmental disabilities.
Genes located in certain “hot spots” mutate much more frequently than those in other parts of the human genome. Evan Eichler, Ph.D., a professor of genome sciences, has been a pioneer in discovering these mutations and detecting their associations with developmental disabilities. “There’s more variation in the genome than was previously thought,” said Eichler, also a research affiliate at the Center on Human Development and Disability. “It’s been a paradigm shift in the field.”
Previous research on inherited developmental disabilities looked at small mutations, known as single nucleotide polymorphisms (SNPs) that can be passed through multiple generations, according to Eichler. However, in recent years Eichler and his colleagues have described large, spontaneous mutations in chromosomal hot spots and linked them to a wide range of developmental disabilities, including intellectual disabilities, epilepsy, and autism spectrum disorders, as well as heart defects, cataracts, and schizophrenia. The mutations are referred to as spontaneous because they can occur in a single generation but not be part of the genetic makeup of a parent.
The newly discovered mutations involve duplications or deletions of large stretches of DNA in areas that are flanked by duplicated sequences. These so-called segmental duplications have emerged recently during the evolution of the genome and carry genes and parts of genes. Segmental duplications make up as much as 5 percent of the human genome.
The flanking duplicated sequences help create mutations called copy number variations (CNV) – gains and losses of DNA. The CNVs described thus far by Eichler and his colleagues are large, about 500,000 or more base pairs of DNA. In contrast, SNPs consist of changes in only a few base pairs.
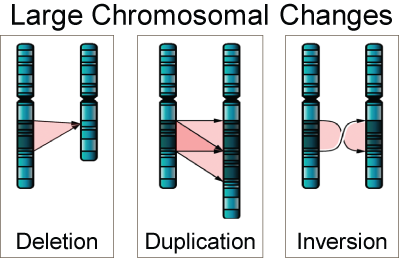
Deletion - DNA not copied, Duplication - DNA copied more than once,
and Inversion- DNA copied in reverse order. All three replication errors
may be involved in causing many developmental disabilities.
The CNVs described by Eichler and his colleagues can occur spontaneously because they happen during meiosis, the type of cell division that produces eggs and sperm. During meiosis, strands of DNA separate and are copied. The copied strands then exchange segments of DNA in a process known as recombination. Recombination allows for the mixing of traits that occurs in families.
Both the process of copying DNA and of recombining can go awry during meiosis, causing mutations in eggs or sperm. “Instead of mom and dad’s chromosomes aligning as they should, they misalign,” said Eichler. The segmental duplications essentially trick the recombination machinery. The enzymes that copy the DNA are confused by the duplicated sequences. As a result, a given sequence may be copied too many times, copied too few times, or skipped altogether. The DNA of an egg or sperm will then differ from that of the parent. For example, according to Eichler, if a sequence is copied too many times, the offspring’s chromosome will be longer than that of the parents, in the same way that individuals with Down syndrome have an extra chromosome compared to their parents.
The same duplicated sequences that cause problems with meiosis can complicate chromosome mapping. Eichler’s group focused on these “difficult bits” while working on the Human Genome Project. In the process of analyzing these areas, they have discovered genes hidden among the segmental duplications. The functions of many of these genes are still totally unknown, said Eichler, but many of them are specific to humans and other closely related primate species.
Variations in the numbers of these newly discovered genes or the genes that are flanked by the duplicated sequences may cause many developmental disabilities associated with the CNVs described by Eichler and his team. “Because these sequences flank a region of the genome that has genes in it, these genes get taken along for the ride,” said Eichler. A CNV may cause a child to receive extra copies, no copy, or a scrambled copy of a given gene. “The child would not simply be the sum of mom and dad, but would be different genetically,” said Eichler. A few years ago, the thought of this much genetic variation occurring in a single generation “was unfathomable,” he said.
Mutations involving CNVs are most likely to occur in certain, very active areas of the genome that have numerous duplicated sequences. The duplications increase the risk of recombination errors that can, in turn, increase the number of duplications. Researchers don’t know why the duplications that feed this cycle initially occurred in these regions. Other primates have similar hot spots in their chromosomes and Eichler and others are studying how CNVs may be linked to human evolution. Regardless of their possible long-term impact on the human species, in the short term CNVs regularly trigger harmful mutations. For example, Eichler and his team have found mutations linked to developmental disabilities in hot spots on chromosomes 1, 15 and 17. These hot spots are “like little volcanoes ready to go off in our genomes,” said Eichler. “It’s scary in a way because it means we’re going to play Russian roulette every time we have a child.”
To better understand the impacts of these hot spots, Eichler and his collaborators have analyzed the genomes of thousands of children with and without developmental disabilities. They have found that the same CNVs can cause a wide range of developmental disabilities. According to research by Eichler and others, the deletion of the same 500,000 base pairs of DNA can result in a child with an intellectual disability or delay, a child with more typical cognitive development who is susceptible to seizures, or a child with more typical development who has a predisposition to develop schizophrenia. Eichler and his team are now working to identify genes within given repeats linked to disability and to then to understand “why increasing or decreasing copies of a gene results in a child that has developmental delay, seizures, epilepsy, or autism.”
“Beyond the diagnostic is the therapeutic,” he said. “Once we understand molecular biology, we may be able to treat it, for example with a medication to mimic the effect of that missing gene. That’s the long-term goal. The baton will be passed onto biochemists, physiologists and neurologists to do that kind of work. Hopefully, downstream, they will come up with therapies that will make a difference in people’s lives in a very meaningful way.”
However, such treatments are decades away. Until then, work by Eichler and his colleagues is allowing better diagnoses of developmental disabilities. Helping parents find diagnoses is very important to Eichler. “I worked with a clinical lab in graduate school, and I heard how most families walked away from a series of genetic tests with no information about the cause of their child’s developmental disability,” he said. Eichler and his colleagues went back to those families after they began to link CNVs to developmental disabilities. With further analyses, Eichler’s team found that many individuals with developmental disabilities in this group of families had one of about 10 specific CNV mutations.
“I can’t tell you how many emails that we’ve had where people now appreciate that they now belong to a group of other families that understand exactly what their kids are going through. That is so rewarding. We’re trying to make a difference in people’s lives, one child and one family at a time,” said Eichler.
|