Pyclaw Solutions¶
All Pyclaw solutions work via the following hierarchy:
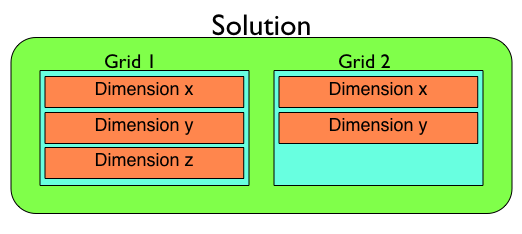
Each solution contains a list of Grids which in turn contain a list of Dimensions, each containing higher level attributes.
pyclaw.solution.Solution¶
- class pyclaw.solution.Solution(*arg, **kargs)¶
Pyclaw grid container class
Input and Output: Input and output of solution objects is handle via the io package. Solution contains the generic methods write(), read() and plot() which then figure out the correct method to call. Please see the io package for the particulars of each format and method and the methods in this class for general input and output information.
Properties : If there is only one grid belonging to this grid, the solution will appear to have many of the attributes assigned to its one grid. Some parameters that have in the past been parameters for all grids are also reachable although Solution does not check to see if these parameters are truly universal.
- Grid Attributes:
‘t’,’meqn’,’mbc’,’q’,’aux’,’capa’,’aux_global’,’dimensions’
Initialization : - The initialization of a Solution can happen on of these ways
- args is empty and an empty Solution is created
- args is a single Grid or list of Grids
- args is a single Dimension or list of Dimensions
- args is a variable number of arguments that describes the location of a file to be read in to initialize the object
- args is a data object with the corresponding data fields
- Input:
- if args == () -> Empty Solution object
- if args == Grids -> Grids are appended to grids list
- if args == Dimensions -> A single Grid with the given Dimensions is created and appended to the grids list
- if args == frame, format=’ascii’,path=’./’,file_prefix=’fort’
- if args == Data, Create a new single grid solution based off of what is in args.
- is_valid()¶
Checks to see if this solution is valid
The Solution checks to make sure it is valid by checking each of its grids. If an invalid grid is found, a message is logged what specifically made this solution invalid.
Output : - (bool) - True if valid, false otherwise
- plot()¶
Plot the solution
- read(frame, path='./', format='ascii', file_prefix=None, read_aux=False, options={})¶
Reads in a Solution object from a file
Reads in and initializes this Solution with the data specified. This function will raise an IOError if it was unsuccessful.
Any format must conform to the following call signiture and return True if the file has been successfully read into the given solution or False otherwise. Options is a dictionary of parameters that each format can specify. See the ascii module for an example.:
read_<format>(solution,path,frame,file_prefix,options={})<format> is the name of the format in question.
Input : - frame - (int) Frame number to be read in
- path - (string) Base path to the files to be read. default = './'
- format - (string) Format of the file, should match on of the modules inside of the io package. default = 'ascii'
- file_prefix - (string) Name prefix in front of all the files, defaults to whatever the format defaults to, e.g. fort for ascii
- options - (dict) Dictionary of optional arguments dependent on the format being read in. default = {}
Output : - (bool) - True if read was successful, False otherwise
- set_all_grids(attr, value, overwrite=True)¶
Sets all member grids attribute ‘attr’ to value
Input : - attr - (string) Attribute name to be set
- value - (id) Value for attribute
- overwrite - (bool) Whether to overwrite the attribute if it already exists. default = True
- write(frame, path='./', format='ascii', file_prefix=None, write_aux=False, options={})¶
Write out a representation of the solution
Writes out a suitable representation of this solution object based on the format requested. The path is built from the optional path and file_prefix arguments. Will raise an IOError if unsuccessful.
Input : - frame - (int) Frame number to append to the file output
- path - (string) Root path, will try and create the path if it does not already exist. default = './'
- format - (string or list of strings) a string or list of strings containing the desired output formats. default = 'ascii'
- file_prefix - (string) Prefix for the file name. Defaults to the particular io modules default.
- write_aux - (book) Write the auxillary array out as well if present. default = False
- options - (dict) Dictionary of optional arguments dependent on which format is being used. default = {}
- aux¶
(ndarray(...,:attr:Grid.maux)) - Grid.aux of base grid
- aux_global¶
(dict) - Grid.aux_global of base grid
- c_center¶
(list) - Grid.c_center of base grid
- c_edge¶
(list) - Grid.c_edge of base grid
- capa¶
(ndarray(...)) - Grid.capa of base grid
- center¶
(list) - Grid.center of base grid
- d¶
(list) - Grid.d of base grid
- dimensions¶
(list) - Grid.dimensions of base grid
- edge¶
(list) - Grid.edge of base grid
- grid¶
(Grid) - Base grid is returned
- grids = None¶
(list) - List of grids in this solution
- lower¶
(list) - Grid.lower of base grid
- maux¶
(int) - Grid.maux of base grid
- mbc¶
(int) - Grid.mbc of base grid
- meqn¶
(int) - Grid.meqn of base grid
- mthbc_lower¶
(list) - Grid.mthbc_lower of base grid
- mthbc_upper¶
(list) - Grid.mthbc_upper of base grid
- n¶
(list) - Grid.n of base grid
- name¶
(list) - Grid.name of base grid
- ndim¶
(int) - Grid.ndim of base grid
- p_center¶
(list) - Grid.p_center of base grid
- p_edge¶
(list) - Grid.p_edge of base grid
- q¶
(ndarray(...,:attr:Grid.meqn)) - Grid.q of base grid
- t¶
(float) - Grid.t of base grid
- units¶
(list) - Grid.units of base grid
- upper¶
(list) - Grid.upper of base grid
pyclaw.solution.Grid¶
- class pyclaw.solution.Grid(dimensions)¶
Basic representation of a single grid in Pyclaw
Dimension information: Each dimension has an associated name with it that can be accessed via that name such as grid.x.n which would access the x dimension’s number of grid cells.
Global Grid information: Each grid has a value for level, gridno, t, mbc, meqn and aux_global. These correspond to global grid traits and determine many of the properties and sizes of the data arrays.
Grid Data: The arrays q, aux and capa have variable extents based on the set of dimensions present and the values of meqn and maux. Note that these are initialy set to None so need to be instantiated. For convenience, the methods emtpy_q(), ones_q(), and zeros_q() for q and emtpy_aux(), ones_aux(), and zeros_aux() for aux are provided to initialize these arrays. The capa array is initially set to all 1.0 and needs to be manually set.
Properties : If the requested property has multiple values, a list will be returned with the corresponding property belonging to the dimensions in order.
Initialization : - Input:
- dimensions - (list of Dimension) Dimensions that are to be associated with this grid
- Output:
- (Grid) Initialized grid object
- add_dimension(dimension)¶
Add the specified dimension to this grid
Input : - dimension - (Dimension) Dimension to be added
- compute_c_center(recompute=False)¶
Calculate the c_center array
This array is computed only when requested and then stored for later use unless the recompute flag is set to True.
Access the resulting computational coodinate array via the corresponding dimensions or via the computational grid properties c_center.
Input : - recompute - (bool) Whether to force a recompute of the arrays
- compute_c_edge(recompute=False)¶
Calculate the c_edge array
This array is computed only when requested and then stored for later use unless the recompute flag is set to True.
Access the resulting computational coodinate array via the corresponding dimensions or via the computational grid properties c_edge.
Input : - recompute - (bool) Whether to force a recompute of the arrays
- compute_p_center(recompute=False)¶
Calculates the p_center array
This array is computed only when requested and then stored for later use unless the recompute flag is set to True (you may want to do this for time dependent mappings).
Access the resulting physical coordinate array via the corresponding dimensions or via the computational grid properties p_center.
Input : - recompute - (bool) Whether to force a recompute of the arrays
- compute_p_edge(recompute=False)¶
Calculates the p_edge array
This array is computed only when requested and then stored for later use unless the recompute flag is set to True (you may want to do this for time dependent mappings).
Access the resulting physical coordinate array via the corresponding dimensions or via the computational grid properties p_edge.
Input : - recompute - (bool) Whether to force a recompute of the arrays
- empty_aux(maux, shape=None, order='C')¶
Initialize aux to empty with given shape
Input : - shape - (tuple) If given, the resulting shape of the auxiliary array will be shape.append(maux). Otherwise it will be (dim.n, maux)
- order - (string) Order of array, must be understood by numpy default = 'C'
- empty_q(order='C')¶
Initialize q to empty
Input : - order - (string) Order of array, must be understood by numpy default = 'C'
- get_dim_attribute(attr)¶
Returns a tuple of all dimensions’ attribute attr
- is_valid()¶
Checks to see if this grid is valid
- The grid is declared valid based on the following criteria:
- q is not None
- All dimensions with boundary conditions set to 0 have valid bc functions
A debug logger message will be sent documenting exactly what was not valid.
Output : - (bool) - True if valid, false otherwise.
- ones_aux(maux, shape=None, order='C')¶
Initialize aux to ones with shape
Input : - shape - (tuple) If given, the resulting shape of the auxiliary array will be shape.append(maux). Otherwise it will be (dim.n, maux)
- order - (string) Order of array, must be understood by numpy default = 'C'
- ones_q(order='C')¶
Initialize q to all ones
Input : - order - (string) Order of array, must be understood by numpy default = 'C'
- qbc()¶
Appends boundary conditions to q
This function returns an array of dimension determined by the mbc attribute. The type of boundary condition set is determined by mthbc_lower and mthbc_upper for the approprate dimension. Valid values for mthbc_lower and mthbc_upper include:
- A user defined boundary condition will be used, the appropriate Dimension method user_bc_lower or user_bc_upper will be called
- Zero-order extrapolation
- Periodic boundary conditions
- Wall boundary conditions, it is assumed that the second equation represents velocity or momentum
Output : - (ndarray(...,meqn)) q array with boundary ghost cells added and set
Note
Note that for user defined boundary conditions, the array sent to the boundary condition has not been rolled.
- remove_dimension(name)¶
Remove the dimension named name
Input : - name - (string) removes the dimension named name
- set_aux_global(data_path)¶
Convenience routine for parsing data files into the aux_global dict
Puts the data from the file at path and puts it into the aux_global dictionary using Data. This assumes that all values in the file are to be put into the aux_global dictionary and the file conforms to that which Data can read.
Input : - data_path - (string) Path to the file to be read in
Note
This will not replace the aux_global dictionary but add to or replace any values already in it.
- zeros_aux(maux, shape=None, order='C')¶
Initialize aux to zeros with shape
Input : - shape - (tuple) If given, the resulting shape of the auxiliary array will be shape.append(maux). Otherwise it will be (dim.n, maux)
- order - (string) Order of array, must be understood by numpy default = 'C'
- zeros_q(order='C')¶
Initialize q to all zeros
Input : - order - (string) Order of array, must be understood by numpy default = 'C'
- aux = None¶
(ndarray(...,maux)) - Auxiliary array for this grid containing per cell information
- aux_global = None¶
(dict) - Dictionary of global values for this grid, default = {}
- c_center¶
(list of ndarray(...)) - List containing the arrays locating the computational locations of cell centers, see compute_c_center() for more info.
- c_edge¶
(list of ndarray(...)) - List containing the arrays locating the computational locations of cell edges, see compute_c_edge() for more info.
- capa = None¶
(ndarray(...)) - Capacity array for this grid, default = 1.0
- center¶
(list) - List of center coordinate arrays
- d¶
(list) - List of computational grid cell widths
- dimensions¶
(list) - List of Dimension objects defining the grid’s extent and resolution
- edge¶
List of edge coordinate arrays
- gridno = None¶
(int) - Grid number of current grid, default = 0
- level = None¶
(int) - AMR level this grid belongs to, default = 1
- lower¶
(list) - Lower coordinate extents of each dimension
- mapc2p = None¶
(func) - Grid mapping function
- maux¶
(int) - Rank of auxiliary array
- mbc = None¶
(int) - Number of ghost cells along the boundaries, default = 2
- meqn = None¶
(int) - Dimension of q array for this grid, default = 1
- mthbc_lower¶
(list) - List of lower bc methods
- mthbc_upper¶
(list) - List of upper bc methods
- n¶
(list) - List of the number of grid cells in each dimension
- name¶
(list) - List of names of each dimension
- ndim¶
(int) - Number of dimensions
- p_center¶
(list of ndarray(...)) - List containing the arrays locating the physical locations of cell centers, see compute_p_center() for more info.
- p_edge¶
(list of ndarray(...)) - List containing the arrays locating the physical locations of cell edges, see compute_p_edge() for more info.
- q = None¶
(ndarray(...,meqn)) - Cell averaged quantity being evolved.
- t = None¶
(float) - Current time represented on this grid, default = 0.0
- units¶
(list) - List of dimension units
- upper¶
(list) - Upper coordinate extends of each dimension
pyclaw.solution.Dimension¶
- class pyclaw.solution.Dimension(*args, **kargs)¶
Basic class representing a dimension of a Grid object
Initialization : - Input:
- name - (string) string Name of dimension
- lower - (float) Lower extent of dimension
- upper - (float) Upper extent of dimension
- n - (int) Number of grid cells
- units - (string) Type of units, used for informational purposes only
- mthbc_lower - (int) Lower boundary condition method to be used
- mthbc_upper - (int) Upper boundary condition method to be used
- user_bc_lower - (func) User defined lower boundary condition
- user_bc_upper - (func) User defined upper boundary condition
- Output:
- (Dimension) - Initialized Dimension object
- qbc_lower(grid, qbc)¶
Apply lower boundary conditions to qbc
Sets the lower coordinate’s ghost cells of qbc depending on what mthbc_lower is. If mthbc_lower = 0 then the user boundary condition specified by user_bc_lower is used. Note that in this case the function user_bc_lower belongs only to this dimension but user_bc_lower could set all user boundary conditions at once with the appropriate calling sequence.
Input : - grid - (Grid) Grid that the dimension belongs to
Input/ouput : - qbc - (ndarray(...,meqn)) Array with added ghost cells which will be set in this routines
- qbc_upper(grid, qbc)¶
Apply upper boundary conditions to qbc
Sets the upper coordinate’s ghost cells of qbc depending on what mthbc_upper is. If mthbc_upper = 0 then the user boundary condition specified by user_bc_upper is used. Note that in this case the function user_bc_upper belongs only to this dimension but user_bc_upper could set all user boundary conditions at once with the appropriate calling sequence.
Input : - grid - (Grid) Grid that the dimension belongs to
Input/ouput : - qbc - (ndarray(...,meqn)) Array with added ghost cells which will be set in this routines
- center¶
(ndarrary(:)) - Location of all grid cell center coordinates for this dimension
- d¶
(float) - Size of an individual, computational grid cell
- edge¶
(ndarrary(:)) - Location of all grid cell edge coordinates for this dimension
- lower = None¶
(float) - Lower computational grid extent
- mthbc_lower¶
(int) - Lower boundary condition fill method. default = 1
- mthbc_upper¶
(int) - Upper boundary condition fill method. default = 1
- n = None¶
(int) - Number of grid cells in this dimension units
- name = None¶
(string) Name of this coordinate dimension (e.g. ‘x’)
- units = None¶
(string) Corresponding physical units of this dimension (e.g. ‘m/s’), default = None
- upper = None¶
(float) - Upper computational grid extent
- user_bc_lower = None¶
(func) - User defined boundary condition function, lower. default = None
- user_bc_upper = None¶
(func) - User defined boundary condition function, upper. default = None
