| 3. |
(a) |
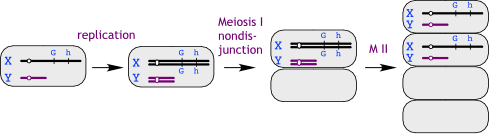
The son's genotype is XhXhY. We don't know whether the extra X chromosome came from his mother or his father**. Assuming that one X came from the mother, she must be carrying at least one copy of the recessive hemophilia allele. So we can list the possible combinations of parental genotypes and then see which one(s) give a probability = 1/2 for an affected child, male or female.
**We are assuming that at least one X came from his mother, although it is possible to imagine complex scenarios where the mother contributed no X and the father contributed both X chromosomes and the Y -- that would require aberrant meioses in the mother and in the father.
| Possible parental genotypes |
The cross |
Predicted outcome |
| XhXh and XHY |
|
Probability of daughter being affected = 0
Probability of son being affected = 1
... this is not the correct set of parental genotypes
|
| XhXh and XhY |
|
Probability of affected child = 1
... this is not the correct set of parental genotypes
|
| XHXh and XHY |
|
Probability of daughter being affected = 0
Probability of son being affected = 1/2
... this is not the correct set of parental genotypes
|
| XHXh and XhY |
|
Probability of daughter being affected = 1/2
Probability of son being affected = 1/2
... this set of parental genotypes fits the data -- the correct parental genotypes are XHXh and XhY
|
(There's also a much shorter way of saying the same thing --
For a son to have a 50% chance of being affected, the mom has to be heterozygous. If she were homozygous dominant, the son would have 0% chance of being affected, as he would inherit the dominant allele from her; if the mom were homozygous recessive, the son would have a 100% chance of being affected. As for the father's genotype, if the father had the dominant allele, his daughter would inherit it and she would have 0% chance of being affected. Therefore, for the daugher to have a chance of being affected, the father must be XhY.
|
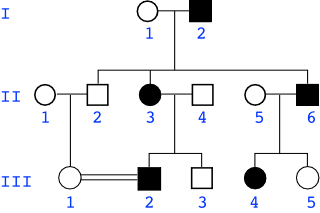 David = II-2
David = II-2
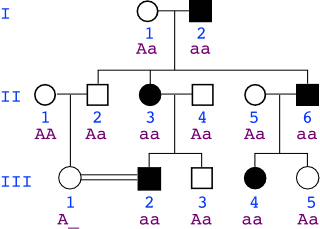 If the trait is autosomal recessive:
If the trait is autosomal recessive:
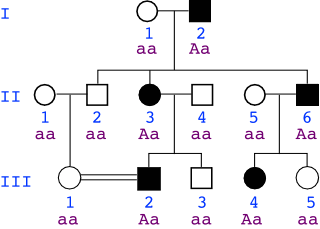 If the trait is autosomal dominant:
If the trait is autosomal dominant: