|
Genetics 371B, Autumn 2000 Problem set 2 -- based on lectures 9-13 |
Due Monday, 30 October at the start of class
| 1. | In a musically inspired moment, you abandon your investigation of pole-vaulting rabbits and switch your research focus, instead, to the genetics of pumpkins. To your supreme gratification, you quickly discover that pumpkins come in two varieties: those that smash readily and those that resist smashing. A cross between smashing- and non-smashing pumpkins gives only non-smashing pumpkins in F1, suggesting that resistance to smashing is dominant while smashing is recessive. To confirm this initial hypothesis, you cross the F1 plants to each other, giving the following F2 progeny:
|
|
| (a) | Does chi-squared analysis support your initial hypothesis that smashing is a simple dominant/recessive trait? Show your work! | |
| (b) | Suggest two reasons why you might have got this deviation from the expected values. | |
| 2. | In fruit flies, speck body (sp), smooth abdomen (sm), and brown eyes (bw) are three recessive traits. Crosses between various strains gave results as follows:
|
|||||||
| (a) | Is any of these three genes located on the X chromosome? Explain in one sentence. | |||||||
| (b) | What two maps for these genes are consistent with the data you have so far? Show your work. | |||||||
| (c) | Outline the cross you would do (using just two of the three genes) to distinguish between the two maps. Explain how your cross would resolve the ambiguity. | |||||||
| (d) | In a cross between a strain that was heterozygous for all three genes and a strain that was fully recessive, the two most abundant progeny phenotypes were wild type and fully recessive, while the two least abundant progeny phenotypes were sm sp and bw. What is the correct order of these three genes on the chromosome? Show your work. | |||||||
| (e) | Interference between the two intervals in this 3-gene region is 0.33. Predict the progeny phenotypes and numbers that you would get if 10,000 progeny were counted in the cross described in (d). | |||||||
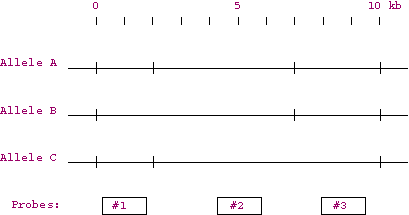
| 3. | Three different alleles of a gene are depicted in the map below. The horizontal line represents the DNA and the small vertical tick marks along the line represent potential cut sites for the restriction enzyme HindIII. Probes 1, 2, and 3 consist of DNA sequences that are identical to the corresponding locations in the map.
|
|
| (a) | Predict the fragment sizes you would get by treatment of each allele with HindIII. | |
| (b) | If you wanted to distinguish between the different alleles in a Southern blot experiment, which of the three probes would you use? Explain. | |
| 4. | The pedigree shows inheritance of an autosomal dominant trait in humans. Also indicated in brackets under each individual is the genotype with respect to a polymorphic microsatellite repeat site (DS-1); the numbers refer to the number of repeats at allele of DS-1.
|
|
| (a) | BRIEFLY explain why this pedigree supports the hypothesis of linkage between the disease gene (G/g) and the polymorphic site DS-1 (i.e., outline what you look for in deciding whether you see evidence of linkage). Identify the putative recombinant progeny. | |
| (b) | Based on these data, what map distance would you assign between the two loci? | |
| (c) | What is the probability of getting this set of progeny genotypes if the two loci are NOT linked? | |