
Since unlinked loci effectively show 50% recombination, as the recombination frequency approaches 50%, the ratio approaches 1.0, and the log of the ratio approaches 0.
|
Genetics 371B, Autumn 2000 Answer key, Problem set 3 |
| 1. | (a) | 
Since unlinked loci effectively show 50% recombination, as the recombination frequency approaches 50%, the ratio approaches 1.0, and the log of the ratio approaches 0. |
| (b) | Curve 3 (PL1-PL2) shows Lod >3 for the range 10-25% recombination. Therefore, there is significant evidence in favor of linkage between the two loci; the distance between the loci is between 10 cM and 25 cM, with a maximum likelihood at 15 cM (the apex of the curve). Furthermore, the curve dips below Lod = -2 | |
| (c) | PL1 and PL2 show significant likelihood of linkage to the disease gene locus, while PL3 shows no evidence of linkage. PL4 shows extremely tight linkage to PL3, so if PL3 is unlnked to D, so is PL4. The probable map distance between D and PL1 is about 5 cM (about 5% recombination), while the D-PL2 distance is about 10 cM. The likely distance between PL1 and PL2 is about 15 cM, which would place these two markers on either side of the disease locus D. Therefore, the map for the five loci can be represented as:
|
| 2. | The logic is as follows: if the chromosome that is trisomic is NOT the one carrying the color gene, then the color gene will behave just as expected for a diploid; we should get a 1:1 ratio of colored and colorless progeny from the testcross. If the trisomic chromosome is the one carrying the color gene, then we'll get a different result:
For P1 parents that are normal with respect to the chromosome carrying the color gene (i.e., genotype rr), the F1 progeny are expected to be Rr and the progeny of the testcross should be colored and colorless in 1:1 ratio. If the P1 parent is trisomic for the chromosome carrying the color gene, then the F1 progeny will be Rrr. For the sake of clarity, the two "r" alleles will be numbred below as r1 and r2 (although they are identical). So the F1 genotype is Rr1r2; depending on how the homologs pair up, the possible gametes are:
in equal proportions. Note that the second and third pair are genotypically identical, but they are two possible outcomes of the meiosis. Since we are only looking at the diploid progeny, any gamete that is disomic can be eliminated (disomic gametes will give trisomic progeny), leaving R and r gametes is 1:2 proportion. Among the ten crosses, the only exception to the 1:1 pattern is when chromosome 9 is trisomic, in which case a 1:2 ratio of colored and colorless progeny are seen. Therefore, the gene for endosperm color must be located on chromosome 9. |
|
| 3. | (a) | 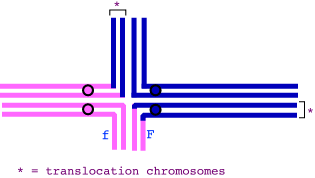 |
| (b) | First, a reminder that a semi-sterile organism is one where half the gametes produced by that individual are inviable. Thus, the translocation heterozygote is semisterile -- the "adjacent" meiotic segregants are inviable, while the "alternate" segregants are viable. Among the "alternate" segregants, the top-right and bottom-left pair (as drawn above) are fully normal and therefore fully fertile; the other pair (top-left, bottom-right) produce progeny that are viable but semisterile again (assuming that the progeny are heterozygous for the translocation too).
If there is no crossing over between the fungus resistance gene and the translocation point, then we should expect equal numbers of normal, sensitive progeny and semisterile, fungus-resistant progeny (i.e., an 'alternate' mode of segregation, with the top-left and bottom-right homologs in the diagram going to one gamete, and the top-right and bottom-left homologs going into one gamete). The progeny are predominantly these phenotypes; the remaining progeny types (normal, resistant, and semisterile, sensitive) must have arisen by recombination between the disease-resistance gene and the translocation point. These two types account for 240 of 2000 progeny, so the map distance between the fungus-resistance gene and the translocation point is 12 cM. Note: if a crossover occurs between the translocation point and the centromere, the resulting recombinant gametes would likely not give viable progeny, as they'd be lacking a portion of a chromosome. (Draw if out if you're unsure!) That's why the resistance locus is depicted as being on the translocated segment. |
| 4. | (a) | The F1 females are fully heterozygous; meiosis in these females should produce recombinant gametes. However, only the parental (NCO) allele combinations are seen, indicating that there has been a chromosomal rearragement between sg and m. Because the males are viable, a deletion is unlikely (although not impossible). Translocations can also be ruled out by the all-or-none nature of the phenotype. Therefore, the rearragement was probably an inversion between sg and m. Recombination between a normal chromatid and an inversion chromatid would lead to acentric and dicentric gametes, both of which would be inviable. The only productive gametes would be the non-recombinant ones. |
| (b) | A deletion of ly+ and tn+ uncovered the recessive alleles. | |
| (c) | Likewise with ly+ and ze+. Since ly is included in both deletions, ly must lie between tn and ze; the gene order is:
|
| 5. | (a) | Green and rough are on one arm of the chromosome, while airborne and fat are on the other arm. Green is associated with single-phenotype and twin-phenotype spots, while rough is associated only with twin-phenotype spots. Therefore, green must be distal to rough (i.e., further from the centromere than rough), such that a single crossover between the centromere and rough gives a twin-phenotype spot, while a crossover between rough and green give a lone green spot. Likewise, fat must be distal to airborne. The meiotic map distances are expected to be proportional to the relative mitotic recombination frequencies, as shown below:
|
||||
| (b) | A double crossover, one crossover between CEN and Rough, and one between Rough and Green, could give lone rough spots (Gr/gr in the diagram). Likewise with airborne, although that should be much more rare. (Only the left arm of the chromosome is shown in the diagram.)
|
|||||