- 5' AGAGGATCACCAGTACCCAG------(CAG)n-------TCCAACACGTGAGTCTGGAC 3'
- 3' TCTCCTAGTGGTCATGGGTC------(GTC)n-------AGGTTGTGCACTCAGACCTG 5'
Primer:
#1) 5'-TCTCCTAGTGGTCATGGGTC-3'
#2) 5'-AGAGGATCACCAGTACCCAG-3' <--- this one...
#3) 5'-GTCCAGACTCACGTGTTGGA-3' <--- and this one
#4) 5'-TCCAACACGTGAGTCTGGAC-3'
#5) 5'-(CAG)7-3'
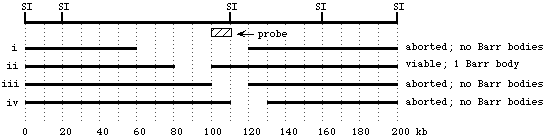
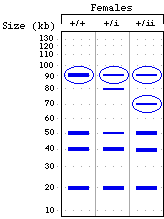
Shown below is the outline of a pedigree for the disease described above, and a representation of a gel showing PCR-amplified fragments detecting the number of CAG repeats. The DNA corresponding to each individual is directly below his or her place in the pedigree. Based on the information you have been given, fill in the pedigree to show the sex and phenotype (affected vs. unaffected) of all individuals except II-9 (save that one for the next question). (10 pts)

The individual appears to have just a single copy of the polymorphic locus, so we'd predict it was an XY male -- except that the allele appears to be derived from the father (26 repeats), and the father should be contributing only a Y to his son.
Several possible explanations:
- non-disjunction in the mom gave an egg with no X chromosome; the sole X chromosome is derived from the dad; the individual is XO female
- the person is an XY male, with a change in one of the alleles from the mom (either the 21-repeat allele changed to 26-repeats, or the >36-repeat allele shrank to 26 repeats)
- the person is an XX female, but a mutation in the primer-binding site prevented the PCR primer from hybridizing to its target -- so that the allele is there, it just couldn't be amplified by PCR
- the person is XX female, but the disease allele expanded even further, to the point that the PCR product was so large that it was outside the range visualized on this gel
- the person is XX {26,26} female -- one of the alleles in the mom changed to a 26-repeat allele; both alleles are present, but since they are the same size, only one band is seen on the gel
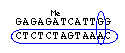
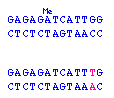
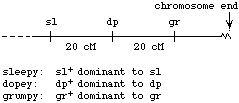
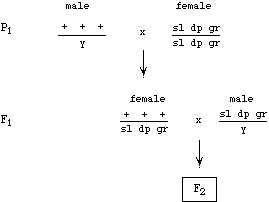
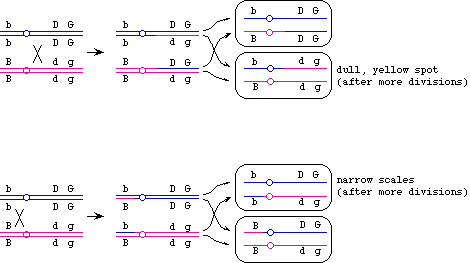
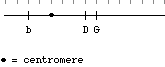 You have a fish that is heterozygous for three scale traits. Broad (B) is dominant to narrow (b), shiny (D) is dominant to dull (d), and green (G) is dominant to yellow (g). The three loci are located on the same chromosome; the arrangement of alleles in the sperm that gave rise to this fish is shown. The tick marks above the map are spaced 5 cM apart.
You have a fish that is heterozygous for three scale traits. Broad (B) is dominant to narrow (b), shiny (D) is dominant to dull (d), and green (G) is dominant to yellow (g). The three loci are located on the same chromosome; the arrangement of alleles in the sperm that gave rise to this fish is shown. The tick marks above the map are spaced 5 cM apart.
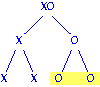 Each egg has an X chromosome; sperm have either
one X or no X. Male progeny result from the fertilization of eggs by sperm
lacking X chromosomes to make XO zygotes. Therefore, the fraction of a male's
progeny that are also male is equal to the fraction of sperm that lack X
chromosomes. For a normal male, half the sperm are O, so half his progeny
will be male. Answer = 0.5
Each egg has an X chromosome; sperm have either
one X or no X. Male progeny result from the fertilization of eggs by sperm
lacking X chromosomes to make XO zygotes. Therefore, the fraction of a male's
progeny that are also male is equal to the fraction of sperm that lack X
chromosomes. For a normal male, half the sperm are O, so half his progeny
will be male. Answer = 0.5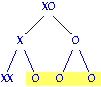 Meiosis II nondisjunction results in three sperm with no X chromosomes and one sperm with 2 X chromosomes. The sperm with no X can fuse with a normal X egg to give an zygote with a normal number of chromosomes, while the sperm with two X's will give an XXX zygote. Therefore, the fraction of gametes that can give progeny with normal chromosome numbers is 0.75 (three out of four). These progeny are expected to be XO males.
Meiosis II nondisjunction results in three sperm with no X chromosomes and one sperm with 2 X chromosomes. The sperm with no X can fuse with a normal X egg to give an zygote with a normal number of chromosomes, while the sperm with two X's will give an XXX zygote. Therefore, the fraction of gametes that can give progeny with normal chromosome numbers is 0.75 (three out of four). These progeny are expected to be XO males.