| 4. |
(22 pts total)
An improbable pair of operons in the mythical bacterium Confusus inhabitabilis is outlined below:

E, F, G, H, J, and K are genes coding for proteins; P and O are the promoter and operator for each operon. G and K are known to be short-lived proteins. The sugar mannose induces transcription of the mannose operon, while melibiose induces transcription of the melibiose operon.
|
|
(a) |
A mutation in G (allele g) causes constitutively low levels of melibiose operon transcription, while a mutation in K (allele k) gives constitutive inactivation of the mannose operon. Explain the mutant phenotypes (specifying gain-of-function vs. loss-of-function) if:
- G is an activator of the melibiose operon and K is a repressor of the mannose operon (4 pts)
g is a loss-of-function mutation of an activator, causing failure of HJK transcription
k is a gain-of-function mutation of a repressor, so EFG transcription is always repressed
- G is a repressor of the melibiose operon and K is an activator of the mannose operon(4 pts)
g is a gain-of-function allele that makes a super-repressor, causing constitutive repression of HJK transcription
k is a loss-of-function mutation of an activator, so EFG transcription does not occur
|
|
|
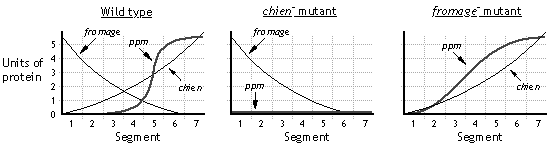
According to Model (ii), K is an activator of EFG transcription; activation of K by mannose induces transcription of EFG. Upon induction of EFG transcription, the level of repressor G increases, so there is increased repression of HJK. The level of activator K therefore drops, and transcription of EFG drops correspondingly. However, in this model, G is a repressor of HJK transcription; melibiose inactivates the repressor, so as long as melibiose is present, the repressor should be inactive, and HJK transcription should occur. Thus, the observation (of transient induction of EFG transcription and sustained induction of HJK transcription) is consistent with model (ii) at least at a simplistic level.
According to Model (i), K is a repressor of EFG transcription; inactivation of K by mannose allows transcription of EFG. In this model, as long as mannose is present, K will remain inactive, allowing continuing transcription of EFG; in contrast, induction of HJK by melibiose should be transient (by the same logic as described above). The observations are not consistent with these predictions.
|
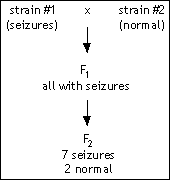 You are studying seizures using a mouse model. You have a true-breeding strain of mice that has frequent, spontaneous seizures. You cross these to another true-breeding strain that does not have seizures. The F1 all have seizures, and a single mating of two F1 animals produces 2 normal mice and 7 mice with seizures.
You are studying seizures using a mouse model. You have a true-breeding strain of mice that has frequent, spontaneous seizures. You cross these to another true-breeding strain that does not have seizures. The F1 all have seizures, and a single mating of two F1 animals produces 2 normal mice and 7 mice with seizures.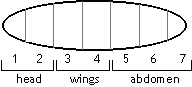 The Great Northwestern Dragonfly develops in a manner similar to Drosophila , with the following difference: while Drosophila has one set of wings and one set of halteres (flight balancers) in the segment immediately posterior to the wing segment, the dragonfly has two sets of wings. The dragonfly larva has seven segments, shown here with their future adult fates.
The Great Northwestern Dragonfly develops in a manner similar to Drosophila , with the following difference: while Drosophila has one set of wings and one set of halteres (flight balancers) in the segment immediately posterior to the wing segment, the dragonfly has two sets of wings. The dragonfly larva has seven segments, shown here with their future adult fates.