| 1. |
(a) |
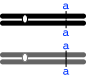
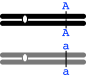
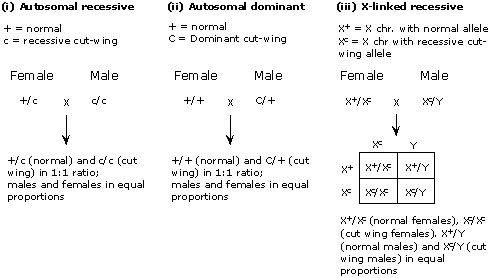
Mode (i) -- autosomal recessive -- will work if the female is heterozygous (+/c) and male is homozygous recessive c/c where "+" denotes the normal allele and "c" denotes the allele for cut-wing--in this case, the recessive allele; the designation "+/c" indicates that the animal is heterozygous--it has the "+" allele on one homolog and the "c" allele on the other). [The female must be heterozygous; it couldn't be homozygous recessive, because it shows the normal phenotype, and it couldn't be homozygous dominant, or the progeny would all be heterozygous and therefore show the dominant phenotype.]
Mode (ii) -- autosomal dominant -- would work also, if the female is homozygous recessive (+/+) and the male is heterozygous (C/+). Note that here, allele C is dominant and causes the cut wing phenotype. Using the same logic as above, the male has to be heterozygous.
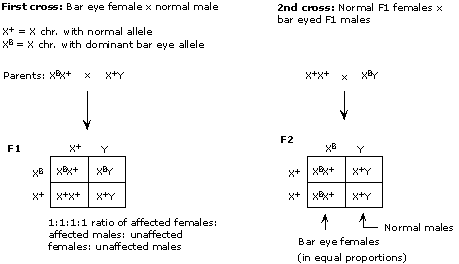
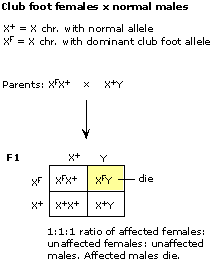
Mode (iii) -- X-linked recessive -- will work if the female is heterozygous and the male carries the recessive, cut-wing allele (c) on his sole X chromosome. The female cannot be homozygous normal because then she would have only normal-winged offspring (heterozygous females, or males with the normal X).
Mode (iv) -- X-linked dominant -- is not possible because if the male carried the dominant allele (C) on his sole X chromosome, all his daughters would inherit that chromosome and would therefore be affected.
The three modes that are consistent with the results are shown below. Note that for (i) and (ii), sex of the animal is independent of the cut wing phenotype, giving males and females in equal proportions for both cut wing and normal phenotypes.
|
|
(b) |
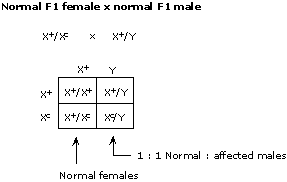
Because males and females from this cross show different phenotype ratios, autosomal modes of inheritance can be ruled out. [To confirm: if the trait were autosomal recessive, the normal x normal cross would be heterozygote x heterozygote, giving a 3:1 ratio of normal : cut wing in the progeny, with males and females in equal numbers; if the cut wing phenotype were autosomal dominant, the normal females and males would all be homozygous normal, as would their progeny.] Therefore, the only mode of inheritance that is consistent with the results is X-linked recessive. The parents and F1 progeny genotypes were as shown in cross (iii) above. The F1 x F1 cross resulting progeny are shown below:
|