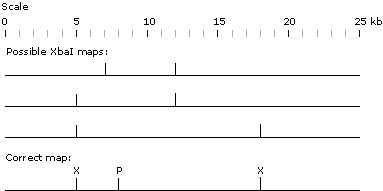
Considering the XbaI digest first: there are three possible ways of explaining this result-- the 5 kb fragment is the middle fragment, the 7 kb fragment is the middle fragment, or the 13 kb fragment is the middle fragment (shown below as the three separate horizontal lines; the bars sticking up from the horizontal lines represent the possible locations of the XbaI sites). Now consider the PstI digest. It cuts only once in the molecule, and it makes an 8 kb fragment and a 17 kb fragment. Therefore, the PstI site has to be 8 kb from one end of the molecule (either the right end or the left end). The double digest resolves the ambiguities. After digesting with both enzymes, we get 4 fragments (as we should); when we compare the XbaI single digest with the double digest, we see that two of the resulting fragments (the 5 kb fragment and the 7 kb fragment) are the same between the two digests. However, instead of the 13 kb fragment, we now gt two fragments, 3 kb and 10 kb--i.e., PstI cuts within the 13 kb fragment produced by XbaI to give a 3 kb fragment and a 10 kb fragment.
Putting these bits of information together--Pst I cuts 8 kb from one end of the DNA molecule; this cut site is within the 13 kb fragment produced by XbaI, and is 3 kb from one end of that 13 kb fragment. The only XbaI map that allows these results is the third one (the 13 kb fragment is the middle fragment). The completed map is shown at the bottom. (If you don't understand this--take each of the three possible XbaI maps, and in each one, try placing the PstI site 8 kb from one end, and see if that gives the expected fragments; then try placing the PstI site 8 kb from the other end and see if that works; etc.)