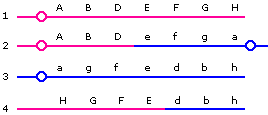
Products 1 and 3 will be viable. Product 2 is dicentric, has two copies of gene A and lacks gene H; product 4 has two copies of gene H, and lacks gene A and a centromere. Therefore, these two products will probably fail to form viable progeny.
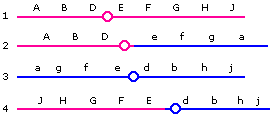
Again, Products 1 and 3 will be viable, while 2 and 4 (having duplications of certain segments and lacking other segments) will each probably be inviable. (Note that the products depicted here are those that would result from a crossover between the E locus and the centromere; the crossover could just as well occur between the D locus and the centromere.)
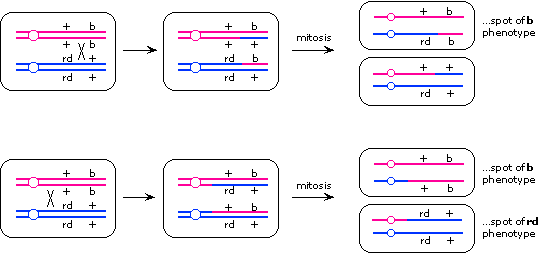
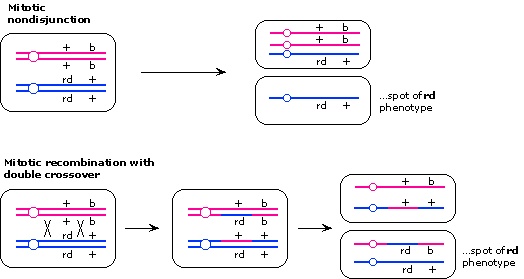
[See Lecture 17 for details on how these results are obtained.]
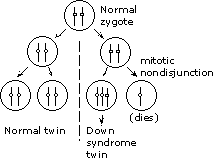
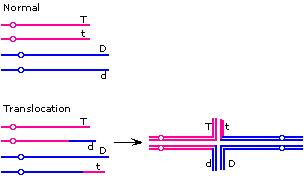

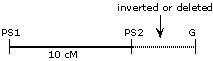 Either a deletion or an inversion in the G-PS2 interval would result in suppression of recombination in that interval, resulting in apparent tight linkage between G and PS2 and an apparent reduction in the PS1-PS2 map distance.
Either a deletion or an inversion in the G-PS2 interval would result in suppression of recombination in that interval, resulting in apparent tight linkage between G and PS2 and an apparent reduction in the PS1-PS2 map distance.