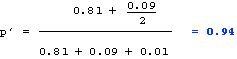Since the alleles are at Hardy-Weinberg frequencies, the frequency of the recessive phenotype = 0.16 = q2 (where q = frequency of allele t). Therefore:
- q = frequency of t = 0.4
- frequency of T = 1 - 0.4 = 0.6
The arithmetic solution to this problem is shown left panel), along with a more general algebraic solution (right panel).
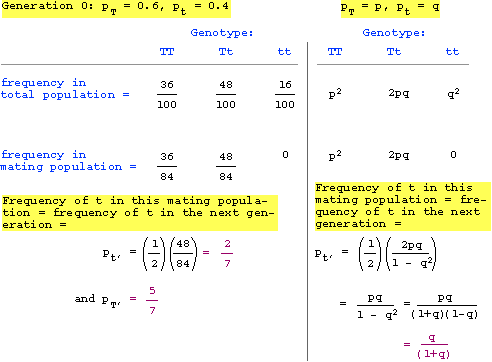
Thus, one generation of restricted mating gives a frequency of T = 5/7 and t = 2/7.
Tn = T0e-µn
- where T0 = initial frequency of T = 0.8
- e = base of natural log
- µ = mutation rate
- n = # of generations = 10000
Therefore, after 10000 generations,
frequency of T = 0.8e-0.02
= 0.784
and the frequency of t = 1 - 0.784 = 0.216
Plugging in the mutation rate, we get N is at least 500,000.