|
Continuing with the map shown in Question 3...
Homozygous {50,50} tissue culture cells in G1 phase of the cell
cycle were exposed to the alkylating agent ethyl methane sulfonate.
In one of those cells the middle Hind III site was alkylated at
the G base indicated by the asterisk:
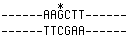
Starting with this DNA, diagram the products after one round of
DNA replication and after two rounds . If you could do a Southern
blot on each of the four cells (after two rounds of cell division)
using Hind III and the same probe as before, what size fragments
would you detect? Assume that only one homologue in the original
cell was modified by EMS. Note that the recognition sequence for
Hind III is 5'-AAGCTT-3'. |
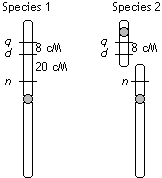 Coin production in a certain species (Species 1) of Money Tree
is controlled by three linked genes: q+ (dominant over q) allows production of quarters; d+ (dominant over d) allows production of dimes; and n+ (dominant over n) allows production of nickels. The recessive condition is lack
of coin production. A linkage map of the region is shown. A closely
related species (Species 2) also has the same genes and alleles,
but here the chromosome has been split in two such that locus
n is on a separate chromosome as shown.
Coin production in a certain species (Species 1) of Money Tree
is controlled by three linked genes: q+ (dominant over q) allows production of quarters; d+ (dominant over d) allows production of dimes; and n+ (dominant over n) allows production of nickels. The recessive condition is lack
of coin production. A linkage map of the region is shown. A closely
related species (Species 2) also has the same genes and alleles,
but here the chromosome has been split in two such that locus
n is on a separate chromosome as shown.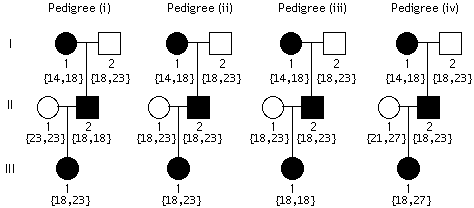
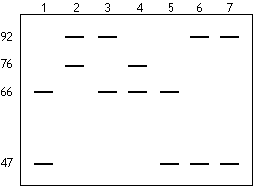 A Southern blot experiment was performed on Hind III-cut DNA obtained
from a woman and her six children. The resulting image is depicted;
the numbers on the side refer to the number of CCTA repeats at
the polymorphic site. Which lane has the mother's DNA? What was
the father's genotype for the polymorphic site? Explain BRIEFLY.
A Southern blot experiment was performed on Hind III-cut DNA obtained
from a woman and her six children. The resulting image is depicted;
the numbers on the side refer to the number of CCTA repeats at
the polymorphic site. Which lane has the mother's DNA? What was
the father's genotype for the polymorphic site? Explain BRIEFLY.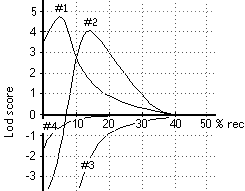 The graph shows LOD score curves for linkage between a disease
gene and four different polymorphic loci (#s 1-4). Interpret the
data for each polymorphic site, stating whether there is significant
evidence for or against linkage. If there is significant evidence,
also state the approximate minimum and/or maximum distance in
cM between the polymorphic site and the disease gene, and give
the most probable map distance if possible.
The graph shows LOD score curves for linkage between a disease
gene and four different polymorphic loci (#s 1-4). Interpret the
data for each polymorphic site, stating whether there is significant
evidence for or against linkage. If there is significant evidence,
also state the approximate minimum and/or maximum distance in
cM between the polymorphic site and the disease gene, and give
the most probable map distance if possible.