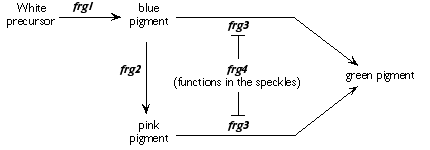
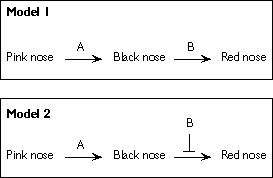
|
In a different population (also showing Hardy-Weinberg frequencies),
20% of the women are heterozygous for red-green colorblindness.
If an affected man marries an unaffected woman of unknown genotype,
what is the probability that their first child will be colorblind? (8 pts)
Note that there can be an affected child only if the mother is
a heterozygote (we know the mom isn't homozygous recessive). And
then it doesn't matter if the child is male or female; the probability
of an affected child is the same for both cases. (If it is a daughter,
she got one colorblindness allele from the dad; whether she is
colorblind depends on which allele she gets from the mom. If it
is a son, he got a Y chromosome from the dad; whether he is colorblind
depends on which allele he gets from the mom.)
Probability that the mom is a heterozygote = 0.2
Probability that she will transmit the colorblindness allele =
0.5
Overall probability of an affected child = (0.2)(0.5) = 0.1.
Note added 12/14/99:
Robert Hennessy points out that the value of heterozygotes = 20%
is for the whole population. Since we know that the mom is not homozygous recessive, we actually need to figure out what fraction
of unaffected women are heterozygotes -- i.e., (# of heterozygotes/#
of unaffected women) rather than (# of heterozygotes/total # of
women). That adjusted fraction will end up being higher than 0.2.
How much higher depends on the allele frequencies, which we don't
have (but can compute). So the answer given above is a rough minimum
estimate rather than the absolute value. Karma points for Robert. |