Chordate DNA-binding zinc fingers
Mammals have an enormously expanded DNA-binding zinc finger superfamily relative to other chordates (for example, humans have upwards of 600 genes compared to a few dozen in fish, frogs, and birds). Though the specific function of most of these genes is mysterious, many specific members are known to be transcriptional regulators, mostly repressors. These facts raise the possibility that this family is a driver of the rapid morphological diversification that characterizes mammalian genera.
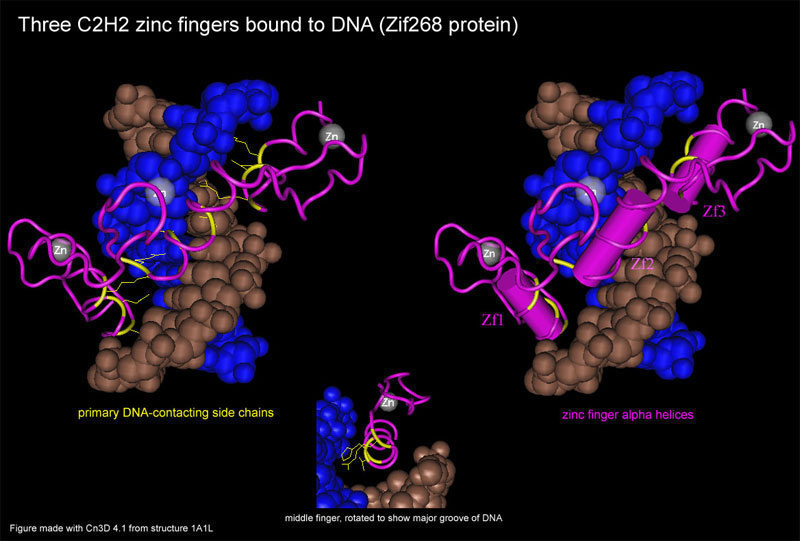
In addition to their potential significance in evolution, DNA-binding zinc finger proteins have a simple modular structure that makes them particularly amenable to computational analysis. Most members of the superfamily consist largely of multiple C2H2 (Kruppel type) zinc fingers. Crystal structures of many such domains are known and the protein residues that contact DNA and determine sequence binding specificity can be predicted from primary amino acid sequence. Each zinc finger repeat has an alpha-helical core that binds to 3 nucleotides in the major groove of DNA. The alpha helix protrudes like a finger from a tetrahedral zinc-binding complex. DNA-binding zinc finger proteins have multiple tandem zinc finger repeats, and the repeats bind to sequential 3 nt blocks in DNA in a beautiful modular structure. Taken together, a series of sequential zinc finger alpa helices run continuously along the major groove of target DNA, much like a series of sausage links running up a helical groove. The zinc-coordinating regions face obliquely away from the DNA and stabilize the structure without making nucleotide-specific contacts. A protein with N zinc finger repeats has the potential to bind a specific DNA sequence of length 3N. Since most members of the superfamily have 8 or more zinc fingers, they are capable of highly specific sequence recognition. Though it is not yet possible to predict DNA-binding specificity from protein sequence alone, the key specificity-determing amino acid residues are well defined and invariantly located with respect to the 2 cysteine (C2) and 2 histidine (H2) zinc-coordinating residues.

In part because the family is so large and complex, my work has thus far focused on the largest specific family within this superfamily - those that contain a Krab transcriptional repressor domain at their N-terminus. About half of the members of the superfamily in humans are in the Krab family.
The Krab C2H2 Zinc Finger Family
A Krab domain is located near the N-terminus of a large family of C2H2 zinc finger proteins, including about 350 in humans. Evidence from many specific cases indicates that the Krab domain confers strong transcriptional repressor activity, with the zinc finger domain conferring sequence-specific DNA binding.
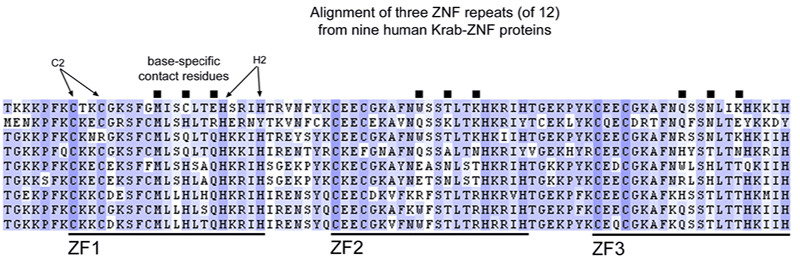
Analysis of molecular evolution among members of the Krab-Zf family reveals extraordinary patterns:
Taken together, these results suggest that Krab-Zf genes frequently duplicate and that the duplicate genes are under strong selective pressure to change their DNA binding specificity during evolution in mammalian lineages. I speculate that this process is a major contributor to the rapid morphological diversification that has long been known to characterize mammalian lineages.
Points 1, 2, and 3 above are apparent on this protein tree (huge image, suggest download to view).
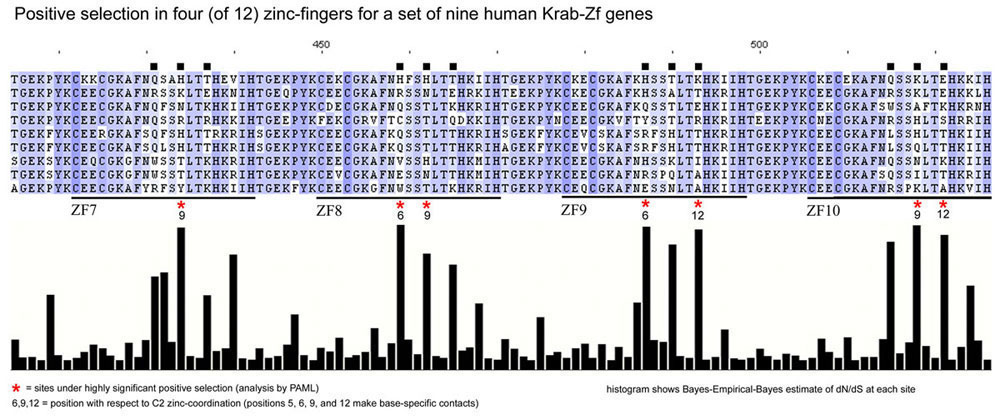
Point 4 above is apparent on the figure below (excerpted from an extensive data set showing similar patterns of positive selection among many genes, usually spread across multiple zinc fingers).
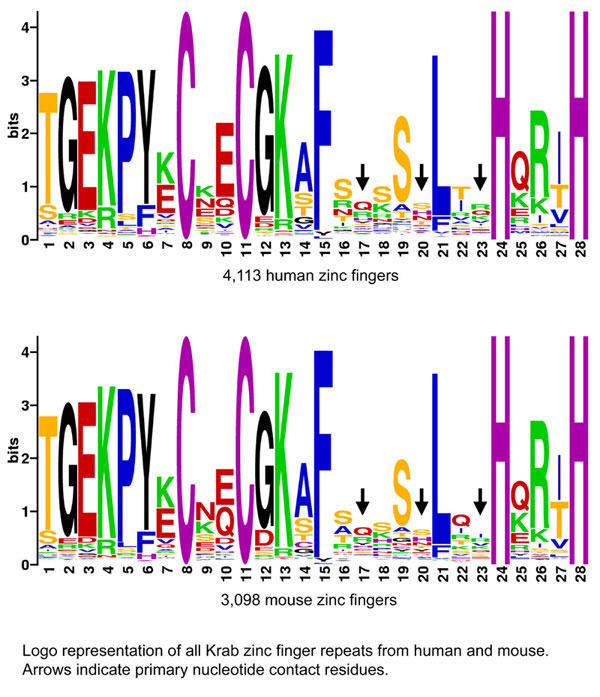
Krab zinc fingers are highly diversified in nucleotide-contacting residues
Another way of viewing the selection for diversity in the nucleotide-contacting residues is shown in the logos below. Each represents all of the zinc fingers from human or mouse Krab-Znf genes aligned (regardless of gene of origin). The human and mouse patterns are nearly identical to each other, and both are characterized by striking amino acid diversity at all three major nucleotide contact residues, embedded in an otherwise conserved zinc finger repeat. When mouse and human fingers are combined, 1,906 of the 8,000 possible amino acid combinations in the three major nucleotide contact residues are represented. The fact that nearly all of these fingers are characterized by highly conserved framework residues suggests the remarkable possibility that most or all of these combinations can function in DNA binding.
A simple explanation of this pattern is that the zinc finger forms a rigid framework that positions the sequence-specificity side chains, an explanation that is supported by various published structural studies.
High conservation of zinc fingers in ortholog pairs
If the sequence diversity in the nucleotide contact residues among expanded Krab-Znf proteins were due to selective pressure to change DNA binding specificity, then one-to-one Krab-Znf orthologs between mouse and human should have highly conserved Znf repeats, particularly in the nucleotide contacting residues. This is precisely what is observed for the 10 clearest ortholog pairs (assigned based on bootstrapped protein trees and synteny). For example, proteins encoded by the ortholog pair ENSG00000198315-ENSMUSG00000063894 align with high quality and have 71 amino acid changes (12.3%), only four of which affect the nine zinc finger repeats (which constitute half of each protein) and none affect the nucleotide contact residues.
These results indicate that evolutionarily stable Krab-Znf genes are characterized by strong purifying selection in the DNA binding domains, reinforcing the significance of the high sequence diversity in unstable Krab-Znf genes.