|
|
Tutorial: Interaction between MakeIndexFile and the PHG
MakeIndexFile
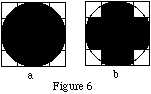
Assume that we have a one slice, 4x4 activity object, with x and y values ranging from —2 to 2 cm. Using MakeIndexFile, assign the activity indexes in two different ways, as follows:
(a) Fill the entire object with an index of 1.
(b) Fill the entire object with an index of 0, then create a cylindrical "object" with an index of 1, centered at (0,0,0), x_radius = y_radius = 2cm, and with z_length covering the full length of the slice. (If the SimSET provided activity index translation file and activity table are used, an index of 0 will correspond to an activity concentration of 0, and an index of 1 will correspond to an activity concentration of 0.000001 Ci/cc.)
MakeIndexFile will fill the objects as follows:
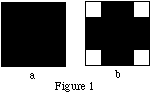 where black represents an index of 1, white an index of 0.
where black represents an index of 1, white an index of 0.
The corner voxels of b are not filled because of the way that MakeIndexFile fills objects: it looks at every voxel to see if the center of the voxel falls within the object. If so, the voxel is assigned the index value for the object, otherwise it is left alone. As can be seen in Figure 2, the centers of the corner voxels do not fall inside the cylinder, but the centers of all other voxels do.
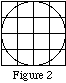
(MakeIndexFile’s one exception to this method of filling objects is when the chosen object is a voxel. Then the user gives the coordinates of any point within the voxel, and the voxel is filled with the given index. Thus, for this example, giving (0.01,0.03,0) as the coordinates of a voxel would cause the following voxel to be filled:
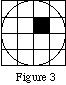 However, giving (0,0,0) at the coordinates of a voxel would lead to an undefined action, as the origin is at the intersection of four voxels.)
However, giving (0,0,0) at the coordinates of a voxel would lead to an undefined action, as the origin is at the intersection of four voxels.)
PHG
 The PHG determines the weight of decays to simulate in a given voxel by multipling the chosen activity concentration times the volume of the voxel, times the scan time, times 3.7E10 decays/Ci/cc. There are three ways af spreading this decay weight around the voxel: uniformly distributed (the default); point source voxels; or line source voxels. If uniform distribution is selected, then the location for each decay is randomly selected within the voxel; if point_source_voxels = true, then every decay occurs at the center point of the voxel; and if line_source_voxels = false, then every decay occurs on the line parallel to the tomograph axis through the center point of the voxel, with the z coordinate randomly sampled within the voxel. Thus, when uniform distributions are selected, the decays are generated from the activity maps given in Figure 1. However, point and line source voxels would be sampled from the following distribution:
The PHG determines the weight of decays to simulate in a given voxel by multipling the chosen activity concentration times the volume of the voxel, times the scan time, times 3.7E10 decays/Ci/cc. There are three ways af spreading this decay weight around the voxel: uniformly distributed (the default); point source voxels; or line source voxels. If uniform distribution is selected, then the location for each decay is randomly selected within the voxel; if point_source_voxels = true, then every decay occurs at the center point of the voxel; and if line_source_voxels = false, then every decay occurs on the line parallel to the tomograph axis through the center point of the voxel, with the z coordinate randomly sampled within the voxel. Thus, when uniform distributions are selected, the decays are generated from the activity maps given in Figure 1. However, point and line source voxels would be sampled from the following distribution:
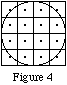
 After a decay is generated, it is checked to see if it occurs within the object cylinder (the largest circular cylinder that fits within the object). If not, it is discarded without be tracked or counted in the PHG statistics. This is equivalent to doing an intersection of the activity object with the following cylinder:
After a decay is generated, it is checked to see if it occurs within the object cylinder (the largest circular cylinder that fits within the object). If not, it is discarded without be tracked or counted in the PHG statistics. This is equivalent to doing an intersection of the activity object with the following cylinder:
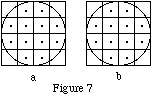
As this occurs after the generation of decay locations, the effective activty maps for the examples in Figure 1 using uniform activity distribution are:
 The activity is uniformly distributed over the portions of voxels contained within the object cylinder.
The activity is uniformly distributed over the portions of voxels contained within the object cylinder.
As all the events in point and line source voxels are concentrated at a single (x,y) coordinate pair, the activity in a voxel is either wholly contained in the object cylinder or wholly outside it. For the activity distributions given in Figure 12, the effective activity distributions end up being identical:
Last revised by: Steven Vannoy Revision date: 2 Feb 2000