Antimicrobial Resistance (AMR) Overview
Antimicrobial resistance (AMR) is a major threat to health and human development, affecting our ability to treat a range of infections. Treatments for a growing number of health care-associated infections (HAI) have become less effective in many parts of the world due to increasing incidence of infections becoming resistant to antimicrobials. However, a large proportion of these infections can be prevented by implementing infection prevention and control (IPC) measures. In this module, you will learn about what AMR is and how resistant infections occur, which pathogens cause the biggest problems globally and in the health care setting, the risk factors and causes of AMR and, most importantly, the role of IPC in reducing AMR.
Learning Objectives
By the end of this module you will be able to:
- describe the mechanisms of antibiotic resistance;
- list important antibiotic-resistant bacteria, including Gram-positive and Gram-negative bacteria, and note key differences;
- explain why the spread of antibiotic resistance is a major threat in all health care facilities worldwide, and why urgent action is needed; and
- explain factors contributing to the emergence and spread of antibiotic-resistant bacteria between health care facilities and communities.
Learning Activities Estimated time:
-
AMR in the NICU (5 min)
Meheret, a 24-year-old pregnant woman, gives birth to preterm (32 weeks) baby boy Nega at a labour ward in a tertiary referral hospital. Nega is immediately transferred to the neonatal intensive care unit (NICU).
Day 4: Nega develops signs and symptoms of acute respiratory distress syndrome (ARDS). A blood culture and umbilical swab are taken, and Nega is started on ceftazidime and vancomycin (two antibiotics).
Day 5: Nega’s condition continues to deteriorate. The lab phones with the news that they have isolated multidrug-resistant Gram-negative bacteria from the blood culture and umbilical swab: Nega has carbapenem-resistant Enterobacteriaceae (CRE). Ceftazidime is changed to gentamicin.
We will talk about Gram stain tests for bacteria later in this module.Carbapenem-resistant Enterobacteriaceae are a family of bacteria that are resistant to a class of antibiotics known as carbapenems. This class of antibiotics are considered the drugs of last resort for these infections.Day 6: Nega’s condition continues to deteriorate. Dr Azeb changes the antibiotic to colistin, which is used when there are no other treatment options available. The final report from the microbiology lab report indicates that Nega has Klebsiella pneumoniae, which is resistant to all antibiotics, including colistin.
Day 7: Nega develops septic shock with multiorgan failure. He dies the next day.
The NICU had other cases of CRE in the past year but no outbreaks. This was the first case of colistin resistance in the NICU.
On a piece of paper or in the box below, list two or three ways Nega could have gotten the organism into his blood (i.e., what is the most likely source). Then click or tap the Compare answer button.
Other ways such bacteria could have been introduced to the hospital environment include:
- patients who acquired the bacteria in the community, but were unaware of this until admitted;
- visitors to the hospital who did not adhere to proper IPC practices while visiting; and
- patients who travelled abroad and sought care in foreign countries.
These types of infections with resistant bacteria are increasing. This upward trend will cause more patients to die from infection, and we will be unable to provide safe delivery of health care.
-
What Is AMR? (5 min)
When an infection caused by a microorganism is no longer treatable by an antibiotic or antiviral, it has developed antimicrobial resistance. Anti means “against,” micro means “small,” and bial refers to life. In general, resistance develops when microorganisms adapt and grow in the presence of the substance used against them (that is, it resists the effects). AMR is a broad term that applies to:
- fungi becoming resistant to antifungals;
- parasites becoming resistant to antiparasitic drugs;
- bacteria becoming resistant to antibiotics; and
- viruses becoming resistant to antivirals.
All classes of microbes can develop resistance.
Antibiotic resistance is a specific term that refers to a subset of AMR; it refers to bacteria becoming resistant to antibiotics.
In the AMR modules, we will focus on antibiotic resistance. However, many of the recommended activities to reduce antibiotic resistance can also be used to combat resistance in other microorganisms that cause fungal, viral and parasitic diseases.
-
How Resistance Occurs (5 min)
How does a microorganism develop resistance? Antibiotic resistance occurs as part of a natural process in which bacteria evolve; it can be slowed but not stopped.
Watch the animation to see how antibiotic resistance happens.

Animation adapted from CDC. Factors contributing to the acceleration of antibiotic resistance are:1
- overuse and misuse of antibiotics in humans and animals;
- health care transmission;
- environmental contamination; and
- suboptimal vaccination.
Types of Resistance
There are two ways bacteria develop resistance to drugs. Click or tap on the tabs to read more about each one.
Intrinsic resistance
Intrinsic resistance is the natural or innate resistance of bacteria to a particular antibiotic; it depends on the properties of bacteria and their mechanisms of action. For example, Gram-negative bacteria are naturally resistant to vancomycin; enterococci are resistant to cephalosporins.
Acquired resistance
Acquired resistance is when bacteria become resistant to an antibiotic to which it was previously susceptible. This can occur either through chromosomal mutation or by plasmid (genetic material) transfer. Bacteria exchange genetic information with each other through conjugation, phage transduction and natural transformation. This is the most dangerous type of resistance that contributes to the overall spread of antibiotic resistance!
-
Which Bacteria Pose the Greatest Threat? (10 min)
Bacteria are identified through Gram staining, and can be split into two groups. Gram-positive bacteria retain a violet stain in their cellular walls, whereas Gram-negative bacteria do not. Gram-negative bacteria can cause serious health care-acquired infections, which have been found to be associated with increased mortality, prolonged hospital stays and higher health care costs. These microorganisms can colonize a patient and cause various types of infection, such as:
Bacteriologist Hans Christian Gram (1853-1938) devised this method of viewing bacteria.- bloodstream infections, especially in immunocompromized patients or patients with indwelling devices;
- gastrointestinal infections after surgery and intra-abdominal sepsis (gut colonization can result in other infections);
- surgical site infections;
- hospital acquired pneumonia (lung infections); and
- urinary tract infections.
Once the gut is colonized with multiresistant Gram-negative organisms, it can result in various infections in patients, depending on the clinical condition.
The WHO has identified 12 bacteria, or priority pathogens, that pose the greatest threat to human health. Click or tap on each tab to see what they are and which antibiotics they are resistant to. In this module, we will focus on the organisms with asterisks.
Priority 1: Critical
Bacteria Gram identification Carbapenem-resistant Acinetobacter baumannii* Negative Carbapenem-resistant Pseudomonas aeruginosa* Negative Carbapenem-resistant and ESBL-producing Enterobacteriaceae* Negative Extended Spectrum Beta-LactamasePriority 2: High
Bacteria Gram identification Vancomycin-resistant Enterococcus (VRE) faecium* Positive Methicillin-resistant Staphylococcus aureus (MRSA)* Positive Clarithromycin-resistant Helicobacter pylori Negative Fluoroquinolone-resistant Campylobacter spp. Negative Fluoroquinolone-resistant Salmonellae Negative Cephalosporin and/or fluoroquinolone-resistant Neisseria gonorrhoeae Negative Priority 3: Medium
Bacteria Gram identification Penicillin—non-susceptible Streptococcus pneumoniae Positive Ampicillin-resistant Haemophilus influenzae Negative Fluoroquinolone-resistant Shigella spp. Negative Let us look at a few pathogens in more detail. Click or tap on each tab to learn more about antibiotic-resistant microorganisms.
Methicillin-resistant S. aureus (MRSA)
Reservoir: S. aureus live in the nose, and in moist and hairy areas of the body (e.g., the groin and axillae). The images below show the average possibility (per 100 people) of S. aureus carriage in the various body sites of healthy adults. Note that the percentages do not match those for MRSA (which are a lot lower), but the images show the body sites where it is most commonly found, both in the general population and in nasal carriers of S. aureus.
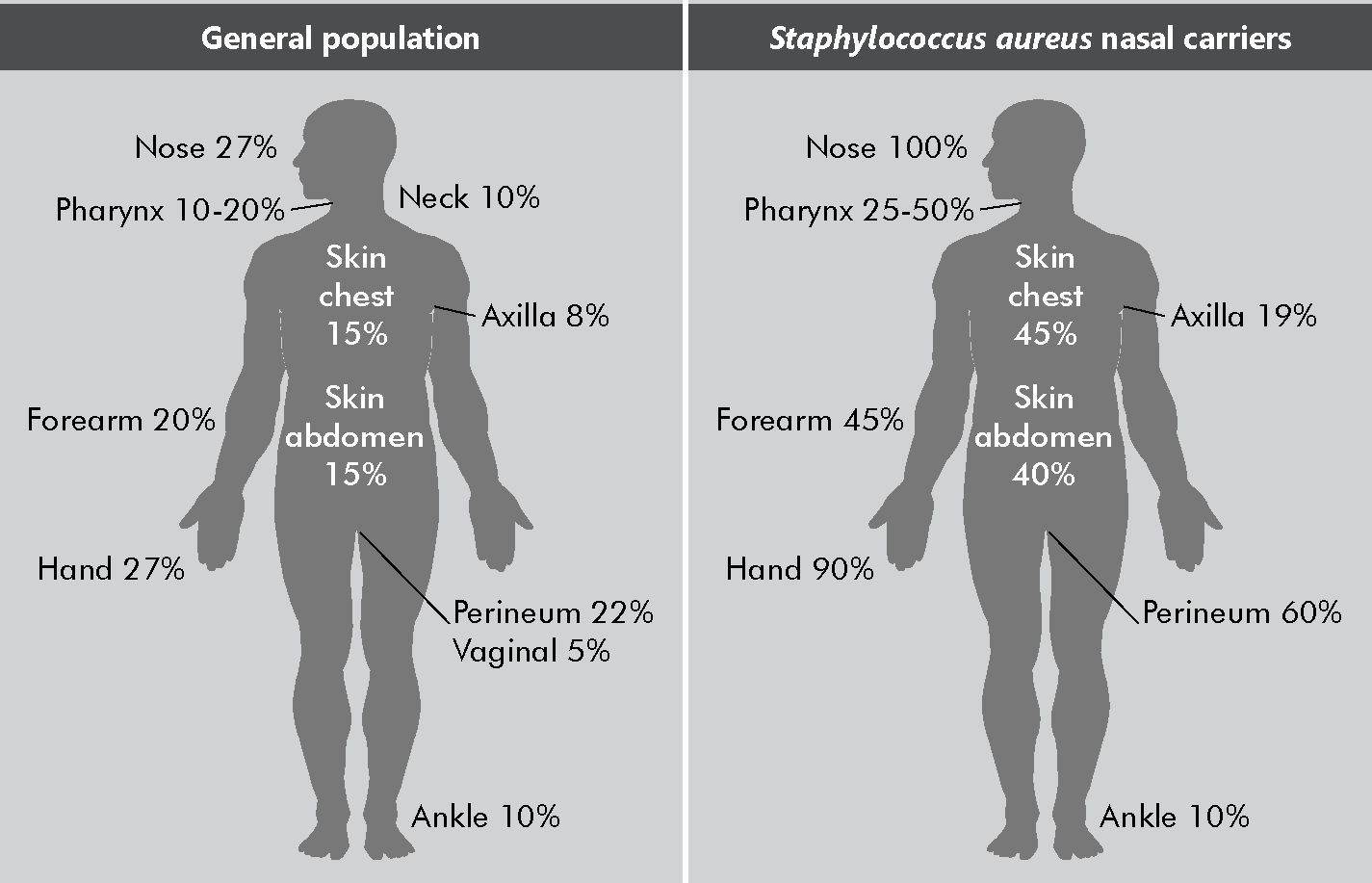 Adapted from Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–62.
Adapted from Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–62.Transmission: Direct and indirect contact (most frequently via hands) and droplets; commonly found in health care settings; highly pathogenic.
Capable of causing disease.Outcome: MRSA is resistant to methicillin/oxacillin/flucloxacillin and cefoxitin. This resistance was first described in 1961, only two years after methicillin was introduced for penicillin-resistant S. aureus. If not controlled, MRSA can achieve continuous presence, or “endemic state,” in a health care facility. Examples of severe infections include skin and soft tissue infection, septic arthritis and osteomyelitis, sinusitis, pneumonia, bloodstream infections, infective endocarditis and food poisoning.
Screening: Swab from nose, axilla and perianal/groin; skin lesions, wounds, incisions, ulcers and exit sites of indwelling devices; newborn umbilicus swab. Routine screening of health care workers (HCWs) is not recommended, but could be considered as part of outbreak control.
Decolonization therapy (to reduce carriage): Feasible in patients who are colonized with MRSA (e.g., using Mupirocin).
Vancomycin-resistant Enterococci (VRE)
Reservoir: Enterococci are opportunistic health care-associated microorganisms—that is, they live in our intestines and skin, usually without causing problems. In specific conditions, however, they can cause disease. They are found in humans and animals, insects and plants, and in the environment. The most prevalent species cultured from humans are Enterococcus faecalis (90%) and Enterococcus faecium (5–10%).
Transmission: Direct and indirect contact (with contaminated objects/equipment, environmental surfaces); commonly found in health care settings with low pathogenicity.
Outcome: Enterococci are resistant to glycopeptides (vancomycin or teicoplanin); for this reason, they are also referred to as GRE. Vancomycin resistance is most common in E. faecium. Infections include urinary tract infection, infective endocarditis, bloodstream infections, surgical site infections, intra-abdominal infections, pelvic infections, and (in rare cases) meningitis and pleural space infections.
Screening: Deep rectal swab, faeces or specimen from colostomy; swab from broken skin (wounds, incisions, ulcers and exit sites of indwelling devices); newborn umbilicus swab.
Decolonization therapy (to reduce carriage): No reliable means for decolonization.
Enterobacteriaceae
Description: A large family of Gram-negative “enteric“ bacteria that includes many harmless symbionts. Disease-causing bacteria in this family include Proteus spp, Enterobacter spp, Serratia spp, Salmonella spp, Shigella spp, Yersinia pestis, Escherichia coli, Klebsiella spp and Citrobacter spp.
Organisms that depend on another type of organism to survive.Reservoir: These types of bacteria are often found in the gastrointestinal tract, but can also be found on the skin, wounds and oropharynx.
Transmission: Direct and indirect contact (most frequently through wounds and stool).
Outcome: They can produce enzymes that determine multiresistance to β-lactam antibiotics such as penicillins, cephalosporins, aztreonam and possibly some carbapenems (ertapenem). For this reason, these bacteria are called extended-spectrum β-lactamases (ESBLs).
Though primarily treated with intravenous carbapenems, these ESBL bacteria (including E. coli, Enterobacter and Klebsiella) can even be resistant to this antibiotic class. These bacteria are known as carbapenem-resistant Enterobacteriaceae (or CRE). Resistance can be caused by several mechanisms, such as:
- active transport of antibiotics out of cells;
- prevention of antibiotics from entering cells; and
- production of enzymes (carbapenemases) disabling the drug molecule.
CRE are treated with intravenous colistin (mostly in combination with other drugs), an antibiotic of last resort. Unfortunately, colistin-resistant CRE was discovered in China in 2015, and found in the UK and US in 2016. Sporadic cases are now regularly reported in most parts of the world.
Screening: Deep rectal swab, faeces or specimen from colostomy; swab from broken skin (wounds, incisions, ulcers and exit sites of indwelling devices); newborn umbilicus swab.
Decolonization therapy (to reduce carriage): Not effective.
Acinetobacter baumannii
Reservoir: Soil, water and sewage, but mostly isolated from hospital environments.
Transmission: Direct and indirect contact (e.g., with contaminated surfaces), water or medication (a common vehicle).
Outcome: Recognized as a significant nosocomial pathogen, especially in critically ill patients in intensive care units (ICUs), and in trauma patients (in wound infections). It is rapidly acquiring resistance to a wide range of antibiotics. Once it is endemic in a hospital, A. baumannii is difficult to eradicate because of its remarkable ability to survive and spread.
Carbapenem resistance among Acinetobacter baumannii (also called carbapenem-resistant Acinetobacter baumannii or CRAB) may be caused by a number of mechanisms. Some strains may be innately resistant to carbapenems, while others contain mobile genetic elements (for example, plasmids, transposons) that result in the production of carbapenemase enzymes (carbapenemases), which break down most β-lactam antibiotics, including carbapenems.
Transposons are sequences of DNA that move (or jump) from one location in the genome to another.Screening: Deep rectal swab, faeces or specimen from colostomy; swab from broken skin (wounds, incisions, ulcers and exit sites of indwelling devices); newborn umbilicus swab.
Decolonization therapy (to reduce carriage): Not effective
Pseudomonas aeruginosa
Reservoir: Soil, water and plants. In the hospital environment, found in hand wash basins and water supplies, especially in high-risk areas (ICU, neonatal care unit, burn unit, etc.).
Transmission: Direct and indirect contact; water is a common vehicle.
Outcome: Responsible for causing a wide variety of infections, particularly in patients with compromised host defence mechanisms (such as those who present with infection in the bloodstream, urinary tract and otitis externa and media), and patients with endocarditis, bacterial keratitis, endophthalmitis and skin infections.
Carbapenem-resistant (CRPsA) may be caused by a number of mechanisms. Some strains may be innately resistant to carbapenems, while others contain mobile genetic elements (for example, plasmids, transposons) that result in the production of carbapenemase enzymes (carbapenemases), which break down most β-lactam antibiotics, including carbapenems.
Screening: Deep rectal swab, faeces or specimen from colostomy; swab from broken skin (wounds, incisions, ulcers and exit sites of indwelling devices); newborn umbilicus swab.
Decolonization therapy (to reduce carriage): Not enough evidence to support recommendation.
-
Knowledge Check (10 min)
-
Why Is Antibiotic Resistance a Problem? (5 min)
Antibiotic resistance has a significant impact not only on patients, but also on the health care facility and system. When bacteria fail to respond to first- or second-line antibiotics, patient morbidity and mortality increase, often resulting in longer hospital stays, thereby placing a greater burden on facilities and the health care system. A study on carbapenemase-producing Enterobacteriaceae found that patients have, on average, 1.79 times higher risk of dying in the ICU than non-colonized patients.2
Antibiotic resistance also increases health care costs: last-line/combination antibiotics are more expensive, patients require more diagnostic tests, and infections result in longer hospital stays and more complications to treat. Ultimately, patients with antibiotic-resistant infections can not only divert resources from regular delivery of care (i.e., because they require additional supplies, isolation, etc.), but also limit the number of available beds for other patients due to increased length of stay.
Resistance also means that there is a limited choice of older, “tried and tested” antibiotics whose efficacy and side effects are well known. Newer antibiotics have restricted licensing conditions due to limited availability of clinical data on their efficacy and significant side effects.
Nearly 40% of the health burden of AMR in European Union countries is caused by infections with bacteria resistant to last-line antibiotics such as carbapenems and colistin.3 Between 2007 and 2015 the burden attributable to infections with K. pneumoniae resistant to carbapenems—a group of last-line antibiotics—increased sixfold. The burden attributable to infections with third-generation cephalosporin-resistant E. coli increased fourfold.
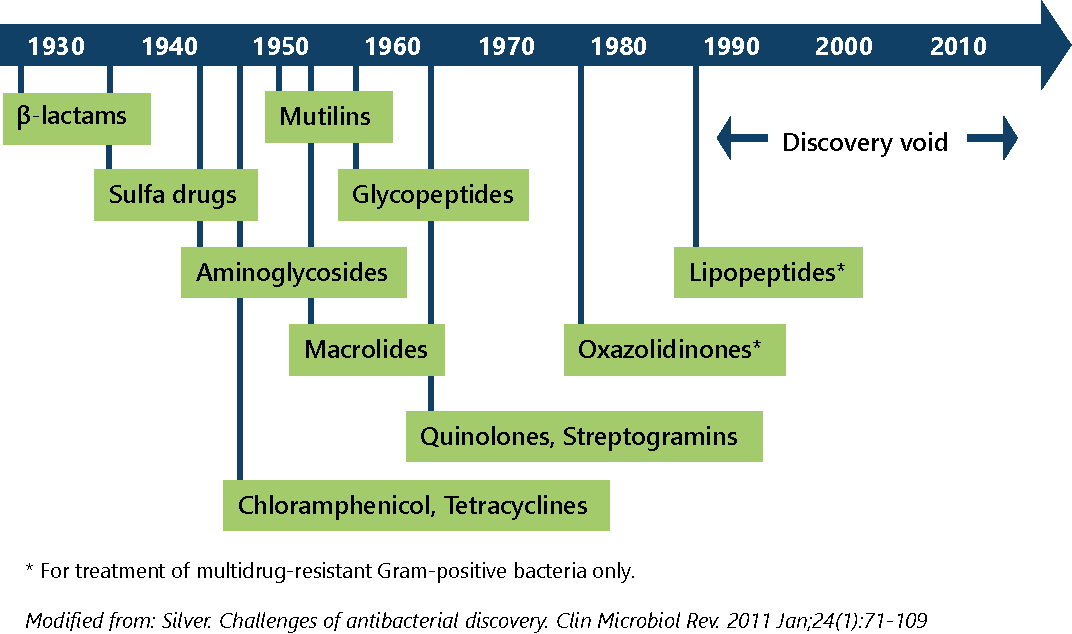
No new registered classes of antibiotics have been discovered since 1987. There is a lack of available antibiotics to treat Gram-negative bacteria.
-
Global Burden of AMR (10 min)
AMR is a global problem and affects most, if not all, countries. Before we discuss the global burden of AMR, take a moment to reflect on these questions:
- Do you know the frequency of antibiotic resistance/AMR in your facility?
- What are the most common health care-associated pathogens in your facility? In your country?
- Do you think there is a difference in rates between high-income countries and low-income countries?
To track the frequency of priority pathogens, WHO developed the Global Antimicrobial Resistance Surveillance System (GLASS). Based on data compiled from patients, laboratories and epidemiology surveillance systems in many countries, WHO has identified bacteria that commonly cause infections in hospitals. WHO also regularly conducts global surveys; one focused on AMR was conducted in 2014 to support activities around Hand Hygiene Day (5 May).
This table shows how many countries provide data on a few of the bacteria that commonly cause infection in hospitals and the community. It is interesting to note that fewer than half the countries report any data on AMR. Among those that do, almost all in WHO regions report more than 50% of notified Enterobacteriaceae are resistant to third-generation cephalosporins, and that more than 50% of notified S. aureus are resistant to methicillin.4
Bacteria commonly causing infections in hospitals and in the community
Name of bacterium/resistance Examples of typical diseases No. out of 194 Member States providing data No of WHO regions with national reports of 50% resistance or more E. coli
- vs 3rd gen. cephalosporins
- vs fluoroquinolones
Urinary tract infections, bloodstream infections 86
92
5/6
5/6
K. pneumoniae
- vs 3rd gen. cephalosporins
- vs carbapenems
Pneumonia, bloodstream infections, urinary tract infections
87
71
6/6
2/6
S. aureus
- vs methicillin “MRSA”
Wound infections, bloodstream infections
85
5/6
In 2014, WHO conducted a laboratory-based global survey on multidrug-resistant organisms (MDROs) in health care. The purpose was to create a snapshot of MDRO prevalence among inpatients in a wide range of health care facilities worldwide, and to collect information about the microbiological methods used for isolation and detection of resistance. Sixty-seven countries and 420 laboratories participated in the survey, which looked at resistance patterns of MRSA, VRE, ESBL-producing Enterobacteriaceae (ESBL-PE), carbapenem-resistant Enterobacteriaceae (CRE) and multiresistant Acinetobacter species (MRAB).5
Survey results showed that the prevalence of ESBL-PE and CRE from blood cultures, and VRE, ESBL-PE and CRE from urine specimens were significantly higher in low- and middle-income countries than in high-income countries.
The Centers for Disease Control and Prevention (CDC) estimated that in 2011 antibiotic resistance caused more than 2 million infections and 23 000 deaths in the USA. At least 500 000 illnesses and 15 000 deaths were from C. difficile infections.6
In the EU/EEA, AMR caused more than 670 000 infections and 33 000 deaths in 2015. These figures represented a doubling in the overall number of infections and a 2.5-fold increase in the number of attributable deaths, respectively, since 2007.3 Resistance to second- and third-line antibiotics is expected to grow by 72% and more than double, respectively, by 2030, compared to 2005.7
-
Risk Factors (5 min)
Let us look at some risk factors that contribute to the emergence and spread of antibiotic-resistant bacteria in health facilities.
On a piece of paper or in the text box below, brainstorm two or three risk factors that contribute to the emergence of antibiotic-resistant bacteria in the health care setting. Click or tap the Compare answer button to learn more.
High-risk hospitals or facility wards may have a high burden of antibiotic-resistant bacteria due to:
- increased use of antibiotics;
- immunocompromized and surgical patients, who are more susceptible to infections;
- increased patient contact due to high intensity of care, resulting in more cross-infection due to breaches in IPC practices; and
- presence of indwelling devices, including intravenous lines, urinary catheters, endotracheal intubation, surgical drains, nasogastric and PEG (gastrostomy and jejunostomy) tubes.
Click or tap the tabs to read about factors that contribute to the spread of antibiotic-resistant bacteria in health care facilities.
System-related factors
- Lack of availability and/or accessibility of up-to-date IPC guidelines.
- Lack of isolation facilities, side wards (especially with ensuite toilets) and facilities to cohort colonised/infected patients.
- Inappropriate/insufficient WASH (water, sanitation and hygiene) infrastructure; inappropriate or hard-to-access materials and equipment related to hand hygiene.
- Lack of good microbiology support/capacity to accurately identify antibiotic-resistant bacteria and appropriate antimicrobial treatments.
- Lack of local surveillance for antibiotic-resistant bacteria.
- Transfers from other healthcare facilities where antibiotic-resistant bacteria are endemic.
- Bed occupancy exceeding the facility’s capacity.
- Increased workloads.
- Inadequate staffing levels.
Healthcare worker-related factors
- Defective IPC practices
- Low or zero compliance with hand hygiene.
- Contaminated environment, items and medical equipment.
- Defective aseptic techniques
Combination of both factors
- Suboptimal or zero implementation of IPC guidelines.
- Lack of identification at the time of admission due to lack of:
- triage and screening of suspected/confirmed patients;
- flagging the notes or charts of patients who are known to be positive for colonization/infection with multidrug-resistant bacteria.
- Failure to isolate suspected/confirmed patients in side rooms with contact precautions.
- Unnecessary movement of patients.
-
Knowledge Check (5 min)
-
Summary (5 min)
In this module, you learnt about what AMR is and how it occurs (intrinsic and acquired). You also learnt about WHO’s priority pathogens—where they occur naturally and what kinds of infections they cause.
The next AMR module will cover IPC strategies (such as hand hygiene) and activities (such as surveillance and antibiotic stewardship) that can help you reduce AMR in your facilities. Remember, AMR is the result of natural adaptation. We will never get rid of AMR entirely, but we can slow it down.
Lindsay Grayson (Australia), “The spread of AMR is just like a bushfire. Yes, we need new firetraps and new helicopters (i.e., new antibiotics), but they’re five or 10 years away. In the meantime, we need a firebreak, and that firebreak is good infection prevention and control.”
-
References
- Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387(10014):176-87. doi: 10.1016/S0140-6736(15)-473-0. Epub 2015 Nov 18.
- Dautzenberg MJ, Wekesa AN, Gniadkowski M, Antoniadou A, Giamarellou H, Petrikkos GL, et al. The association between colonization with carbapenemase-producing enterobacteriaceae and overall ICU mortality: an observational cohort study. Crit Care Med. 2015 Jun;43(6):1170-7. doi: 10.1097/CCM.0000000000001028.
- Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019 Jan;19(1):56-66. doi: 10.1016/S1473-3099(18)30605-4. Epub 2018 Nov 5.
- WHO Antimicrobial Resistance Surveillance System (GLASS). https://www.who.int/glass/en/
- Allegranzi B, Damani N, Gayet-Ageron A, Stewardson A, Wallace S, Pittet D. World Health Organization period prevalence survey on multidrug-resistant microorganisms in healthcare. Vienna, Austria: European Congress of Clinical Microbiology and Infectious Diseases; 2017.
- Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States. Atlanta, GA: US Department of Health and Human Services, CDC; 2013.
- OECD. Stemming the Superbug Tide: Just a Few Dollars More. Paris: OECD Health Policy Studies, OECD Publishing; 2018. Available at oe.cd/amr-2018.
