A certain X-linked dominant disease in humans occurs when there are more than 40 repeats of a CGG trinucleotide in a particular gene. The unique sequence outside the repeat region is as follows:
- 5' CAGTATGCA------(CGG)n-------ATGCGTAAT 3'
- 3' GTCATACGT------(GCC)n-------TACGCATTA 5'
In the lab freezer, you find 5 primers of different sequences (listed below). If you wanted to use PCR to amplify the trinculeotide repeat region, which of these primers would you use?
- Primer:
- #1) 5'-GTCATACGT-3'
- #2) 5'-ATTACGCAT-3' -- "Reverse primer" primes leftward extension from the right end
- #3) 5'-ATGCGTAAT-3'
- #4) 5'-CAGTATGCA-3' -- "Forward primer" primes rightward extension from the left end
- #5) 5'-TGCATACTG-3'
- #1) 5'-GTCATACGT-3'
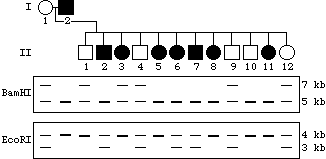
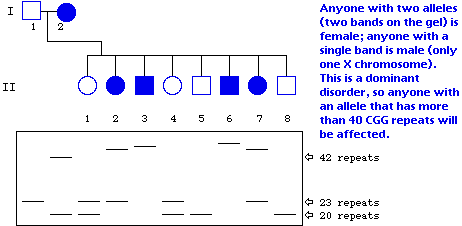
Shown below are the outline of a pedigree for the disease described above, and a representation of a gel showing PCR-amplified fragments detecting the number of CGG repeats. The DNA corresponding to each individual is directly below his or her place in the pedigree. Based on the information you have been given, fill in the pedigree to show the sex of the individual as well as the phenotype (affected vs. unaffected).
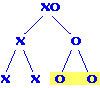 Each egg has an X chromosome; sperm have either one X or no X.
Male progeny result from the fertilization of eggs by sperm lacking
X chromosomes to make XO zygotes. Therefore, the fraction of a
male's progeny that are also male is equal to the fraction of
sperm that lack X chromosomes. For a normal male, half the sperm
are O, so half his progeny will be male. Answer = 0.5
Each egg has an X chromosome; sperm have either one X or no X.
Male progeny result from the fertilization of eggs by sperm lacking
X chromosomes to make XO zygotes. Therefore, the fraction of a
male's progeny that are also male is equal to the fraction of
sperm that lack X chromosomes. For a normal male, half the sperm
are O, so half his progeny will be male. Answer = 0.5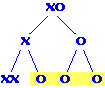 Meiosis II nondisjunction results in three sperm with no X chromosomes,
so the fraction of male progeny goes up to 0.75 (three out of four).
Meiosis II nondisjunction results in three sperm with no X chromosomes,
so the fraction of male progeny goes up to 0.75 (three out of four).