Difference between revisions of "Anatomy Pipeline"
(→AC-PC Aligned Nifti Image) |
(→Data Organization/Naming Convention) |
||
| (29 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
We collect a high resolution T1-weighted image on every subject, and use this image to define the coordinate space for all subsequent analyses. This section describes the processing steps for a subject's T1-weighted anatomy and should be performed before analyzing the rest of their MRI data. | We collect a high resolution T1-weighted image on every subject, and use this image to define the coordinate space for all subsequent analyses. This section describes the processing steps for a subject's T1-weighted anatomy and should be performed before analyzing the rest of their MRI data. | ||
| + | ==Data Organization/Naming Convention== | ||
| + | Step 1: Create a subject ID folder within the anatomy directory | ||
| + | NLR_###_FL_MR# | ||
| + | For Longitudinal scans, you will want to create a folder for each individual scan, as well as a general directory NLR_###_FL to store an AC-PC aligned image of the total average across scans.<br> | ||
| + | [subid] --seen below-- will include the _MR# for each individual scan. | ||
| + | |||
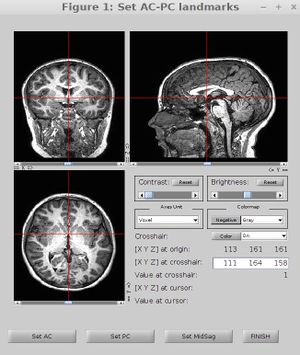
==AC-PC Aligned Nifti Image== | ==AC-PC Aligned Nifti Image== | ||
| − | Data can come off the scanner with arbitrary header information and in parrec format. So for each subject we start by defining a coordinate frame where 0,0,0 is at the anterior commissure, the anterior and posterior commissure are in the same X and Z planes, and the mid-line is centered in the image. Bob Dougherty wrote a nice tool to help with this. See [https://github.com/vistalab/vistasoft/blob/master/mrAnatomy/VolumeUtilities/mrAnatAverageAcpcNifti.m mrAnatAverageAcpcNifti]. The subject's T1-weighted image should be ac-pc aligned, resliced (preserving its resolution), and saved in the subject's anatomy directory. | + | [[File:Ac-pc.jpg|300px|right]] |
| − | cd / | + | Data can come off the scanner with arbitrary header information and in parrec format. So for each subject we start by defining a coordinate frame where 0,0,0 is at the anterior commissure, the anterior and posterior commissure are in the same X and Z planes, and the mid-line is centered in the image. Bob Dougherty wrote a nice tool to help with this. See [https://github.com/vistalab/vistasoft/blob/master/mrAnatomy/VolumeUtilities/mrAnatAverageAcpcNifti.m mrAnatAverageAcpcNifti]. The subject's T1-weighted image should be ac-pc aligned, resliced (preserving its resolution), and saved in the subject's anatomy directory.<br> |
| − | parrec2nii -c -b | + | Step 1: In a terminal, convert the PAR/REC files to nifti images |
| − | im = niftiRead( | + | cd /mnt/diskArray/projects/MRI/[subid] |
| − | mrAnatAverageAcpcNifti({ | + | parrec2nii -c -b *.PAR |
| + | Step 2: In MATLAB compute the root mean squared (RMS) image and then ac-pc align. If a subject has multiple images then the rms operation should be run on each image and then a cell-array with paths to all the images can be pushed through mrAnatAverageAcpcNifti resulting in a very nice anatomy | ||
| + | T1path = 'Path to t1 weighted image'; | ||
| + | T1path = mri_rms(T1path); % Root mean squared image | ||
| + | im = niftiRead(T1path); % Read root mean squared image | ||
| + | voxres = diag(im.qto_xyz)'; % Get the voxel resolution of the image (mm) | ||
| + | mrAnatAverageAcpcNifti({T1path}, '/mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz', [], voxres(1:3)') | ||
==Freesurfer Segmentation== | ==Freesurfer Segmentation== | ||
Freesurfer is a useful tool for segmenting a T1-weighted image and building a cortical mesh. To segment the subject's T1-weighted image using freesurfer from the command line type: | Freesurfer is a useful tool for segmenting a T1-weighted image and building a cortical mesh. To segment the subject's T1-weighted image using freesurfer from the command line type: | ||
| − | recon-all -i / | + | recon-all -i /mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz -subjid [subid] -all |
Or even better use this handy matlab function written by Jon Winawer to run freesurfer and then also build some useful files that we like to use for data visualization such as a high resolution gray/white segmentation. | Or even better use this handy matlab function written by Jon Winawer to run freesurfer and then also build some useful files that we like to use for data visualization such as a high resolution gray/white segmentation. | ||
| − | fs_autosegmentToITK([subid], '/ | + | fs_autosegmentToITK([subid], '/mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz') |
| − | + | For subjects with multiple scans, the output destination should be [subid]_MR[#]. For example, a subject's (205_AB) second scan for the NLR study would be written: NLR_205_AB_MR2. This is in line with the Freesurfer system of notation. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | This | + | |
| − | + | ||
Latest revision as of 22:27, 3 December 2015
Contents
We collect a high resolution T1-weighted image on every subject, and use this image to define the coordinate space for all subsequent analyses. This section describes the processing steps for a subject's T1-weighted anatomy and should be performed before analyzing the rest of their MRI data.
Data Organization/Naming Convention
Step 1: Create a subject ID folder within the anatomy directory
NLR_###_FL_MR#
For Longitudinal scans, you will want to create a folder for each individual scan, as well as a general directory NLR_###_FL to store an AC-PC aligned image of the total average across scans.
[subid] --seen below-- will include the _MR# for each individual scan.
AC-PC Aligned Nifti Image
Data can come off the scanner with arbitrary header information and in parrec format. So for each subject we start by defining a coordinate frame where 0,0,0 is at the anterior commissure, the anterior and posterior commissure are in the same X and Z planes, and the mid-line is centered in the image. Bob Dougherty wrote a nice tool to help with this. See mrAnatAverageAcpcNifti. The subject's T1-weighted image should be ac-pc aligned, resliced (preserving its resolution), and saved in the subject's anatomy directory.
Step 1: In a terminal, convert the PAR/REC files to nifti images
cd /mnt/diskArray/projects/MRI/[subid] parrec2nii -c -b *.PAR
Step 2: In MATLAB compute the root mean squared (RMS) image and then ac-pc align. If a subject has multiple images then the rms operation should be run on each image and then a cell-array with paths to all the images can be pushed through mrAnatAverageAcpcNifti resulting in a very nice anatomy
T1path = 'Path to t1 weighted image';
T1path = mri_rms(T1path); % Root mean squared image
im = niftiRead(T1path); % Read root mean squared image
voxres = diag(im.qto_xyz)'; % Get the voxel resolution of the image (mm)
mrAnatAverageAcpcNifti({T1path}, '/mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz', [], voxres(1:3)')
Freesurfer Segmentation
Freesurfer is a useful tool for segmenting a T1-weighted image and building a cortical mesh. To segment the subject's T1-weighted image using freesurfer from the command line type:
recon-all -i /mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz -subjid [subid] -all
Or even better use this handy matlab function written by Jon Winawer to run freesurfer and then also build some useful files that we like to use for data visualization such as a high resolution gray/white segmentation.
fs_autosegmentToITK([subid], '/mnt/diskArray/projects/anatomy/[subid]/t1_acpc.nii.gz')
For subjects with multiple scans, the output destination should be [subid]_MR[#]. For example, a subject's (205_AB) second scan for the NLR study would be written: NLR_205_AB_MR2. This is in line with the Freesurfer system of notation.