FMRI
From Brain Development & Education Lab Wiki
Contents
Organize the subject's data
Put raw data into:
subjid/date/raw
Hereafter referred to as the session directory Make event files (parfiles) and put the in
subjid/date/Stimuli/parfiles
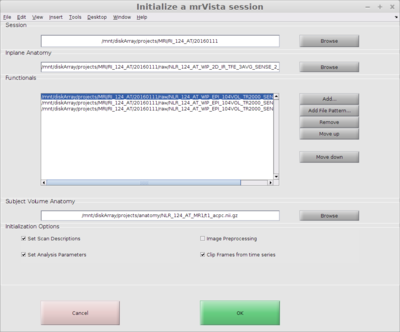
Initialize an mrVista session
In a MATLAB terminal change to the session directory and open mrInit
mrInit
Add files to mrVista session
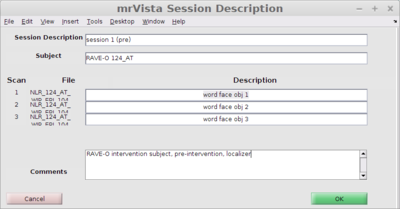
Fill in the session description
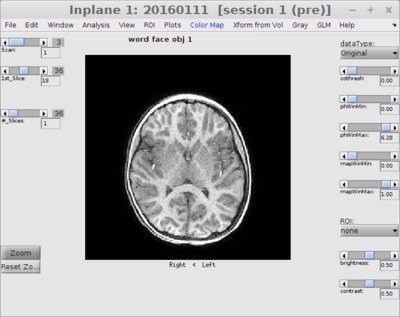
Open Inplane view
mrVista
And run motion correction
Analysis -> Motion Compensation -> Within + Between Scan
Fit GLM
First associate a parfile with each scan and then group all the MotionComp scans so that the GLM is fit to all of them
GLM -> Assign Parfiles to Scan GLM -> Grouping -> Group Scans GLM -> Apply CLM/Contrast New Code
In the GLM window that comes up set the HRF to SPM Difference of gammas and set the number of TRs (in our case 8) and detrend option to Quadratic.