Difference between revisions of "MEG Data Acquisition"
From Brain Development & Education Lab Wiki
| Line 2: | Line 2: | ||
cd /home/jyeatman/git/mnefun/bin | cd /home/jyeatman/git/mnefun/bin | ||
python run_mne_bem.py --subject NLR_201_GS --layers 1 --overwrite | python run_mne_bem.py --subject NLR_201_GS --layers 1 --overwrite | ||
| + | |||
| + | 2. Coordinate alignment. | ||
| + | mne_analyze | ||
| + | File -> Load Surface -> Select Inflated | ||
| + | File -> Load Digitizer Data -> sss_fif -> select any raw file | ||
| + | Adjust -> Coordinate Alignment | ||
| + | View -> Show viewer | ||
| + | Click Options -> check Digitizer Data and HPI and landmarks only, | ||
| + | Click each Fiducial location and then click "Align using fiducials" | ||
| + | Save Default | ||
| + | Make a "trans" folder within the subject's directory | ||
| + | Move transform file and rename subj-trans.fif | ||
2. To visualize source localized data | 2. To visualize source localized data | ||
| Line 11: | Line 23: | ||
[[File:LoadInverse-mne analyze.png]] | [[File:LoadInverse-mne analyze.png]] | ||
| + | To adjust sensor plots: Adjust -> Scales | ||
| + | To adjust source visualization: Adjust -> Estimates | ||
Revision as of 19:49, 23 October 2015
1. After running freesurfer on a subject's T1 anatomy we next need to create a BEM model.
cd /home/jyeatman/git/mnefun/bin python run_mne_bem.py --subject NLR_201_GS --layers 1 --overwrite
2. Coordinate alignment.
mne_analyze
File -> Load Surface -> Select Inflated File -> Load Digitizer Data -> sss_fif -> select any raw file Adjust -> Coordinate Alignment View -> Show viewer Click Options -> check Digitizer Data and HPI and landmarks only, Click each Fiducial location and then click "Align using fiducials" Save Default Make a "trans" folder within the subject's directory Move transform file and rename subj-trans.fif
2. To visualize source localized data
mne_analyze
3. File -> Load Surface -> Select Inflated
4. File-> Open
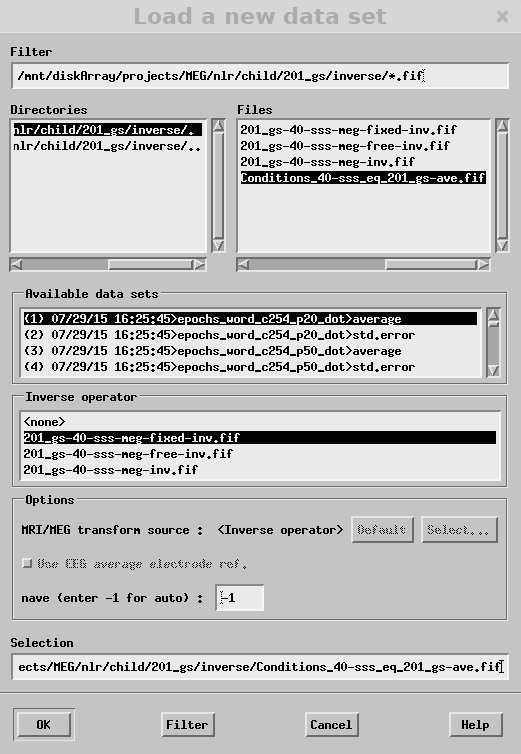 To adjust sensor plots: Adjust -> Scales
To adjust source visualization: Adjust -> Estimates
To adjust sensor plots: Adjust -> Scales
To adjust source visualization: Adjust -> Estimates