MEG Data Acquisition
Creating a BEM model for source localization
After running freesurfer on a subject's T1 anatomy we next need to create a BEM model.
cd /home/jyeatman/git/mnefun/bin python run_mne_bem.py --subject NLR_201_GS --layers 1 --overwrite
Aligning MEG sensor data to the BEM model
Open mne_analyze to compute the coordinate alignment. In mne_analyze load the subject's surface and digitizer data:
File -> Load Surface -> Select Inflated File -> Load Digitizer Data -> sss_fif -> select any raw file
Next adjust how these digitized points align with the scalp in the MRI
Adjust -> Coordinate Alignment View -> Show viewerWithin the viewer window click the "Options" button. Within the "Viewer Options" dialogue check "digitizer data" and "HPI and landmarks only".
Next mark the fiducial locations on the scalp. To do this click LAP, RAP and Nasion in the "Adjust coordinate alignment" dialogue and then mark each spot by clicking on the scalp surface . After marking each location click "Align using fiducials". 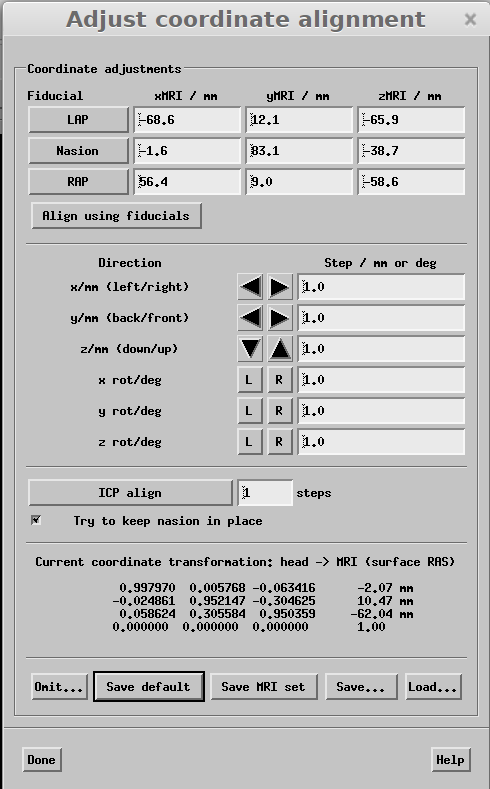
Once you have aligned everything click "Save Default" in the coodinate adjustment dialogue. This will save out the transform in the subjects folder. Then, make a "trans" folder within the subject's directory and move transform file and rename subj-trans.fif
== Visualizing data in source space
mne_analyze File -> Load Surface -> Select Inflated File-> Open
To adjust sensor plots: Adjust -> Scales To adjust source visualization: Adjust -> Estimates