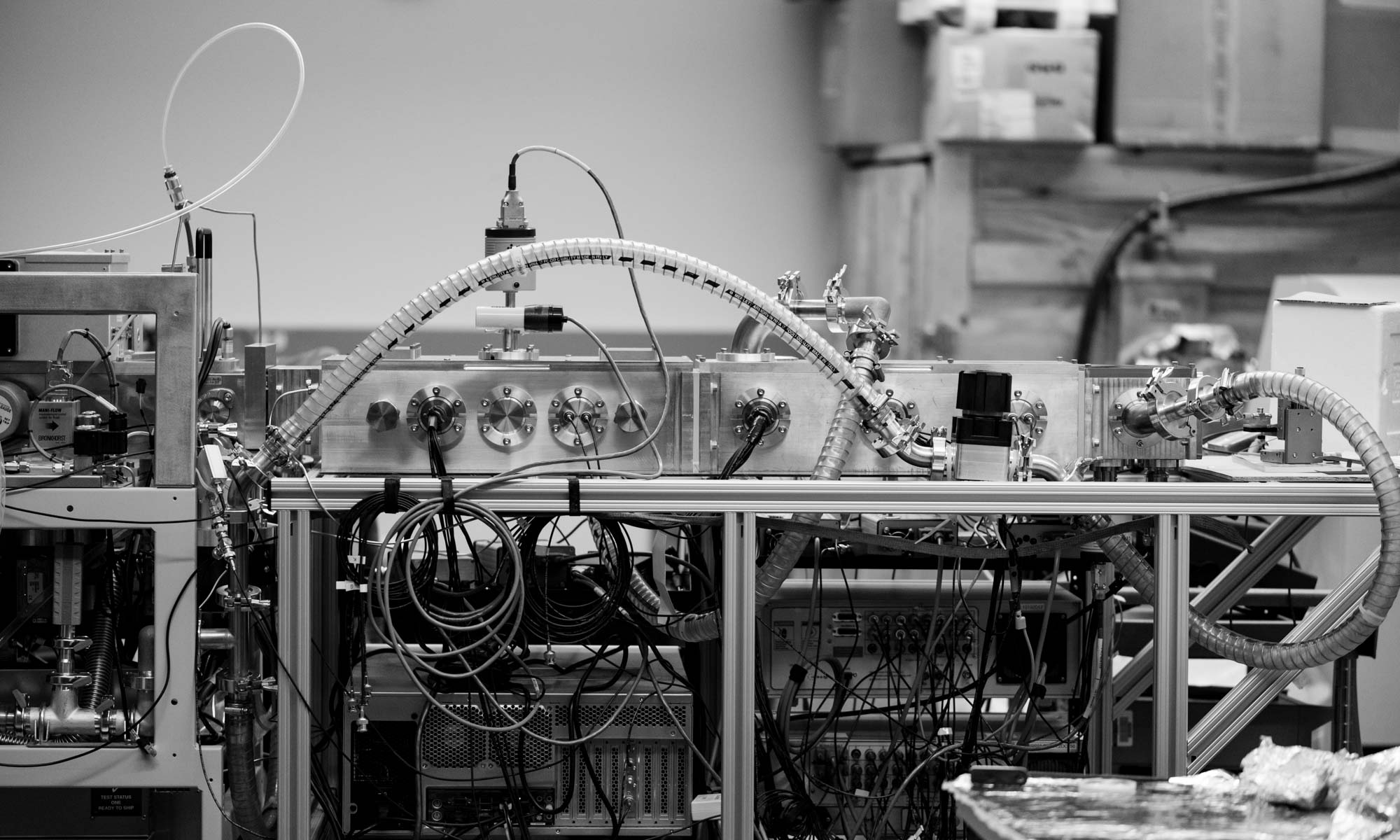
The Bush Lab is a research group in the Department of Chemistry and the Biological Physics, Structure & Design Program at the University of Washington. Our research focuses on the development and application of mass spectrometry and ion mobility spectrometry techniques to elucidate the structures and assembly of protein complexes and subcellular machines.
- Interested in joining the Bush Lab? Click here to learn about current opportunities.
- Our weekly group meetings are held each Friday at 9:30 AM.