![]()
 Animation showing fundamentals of nanopore experiments. As DNA translocates (top), the ion current is blocked (bottom).
Animation showing fundamentals of nanopore experiments. As DNA translocates (top), the ion current is blocked (bottom).
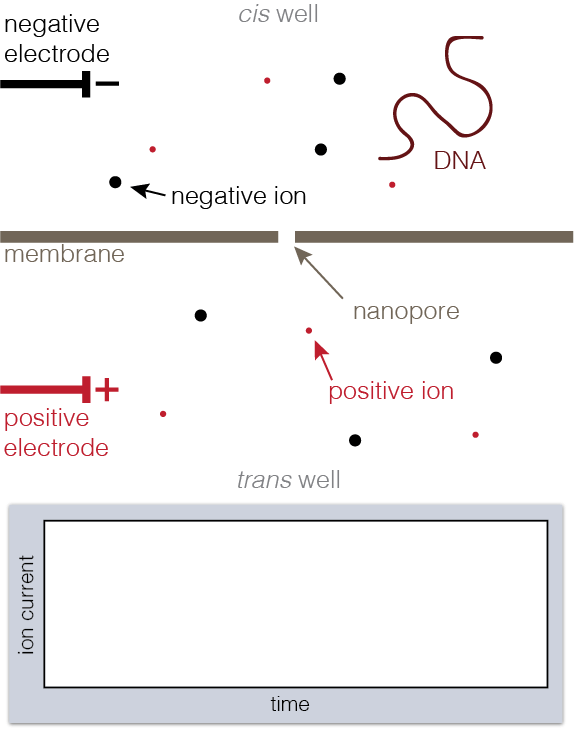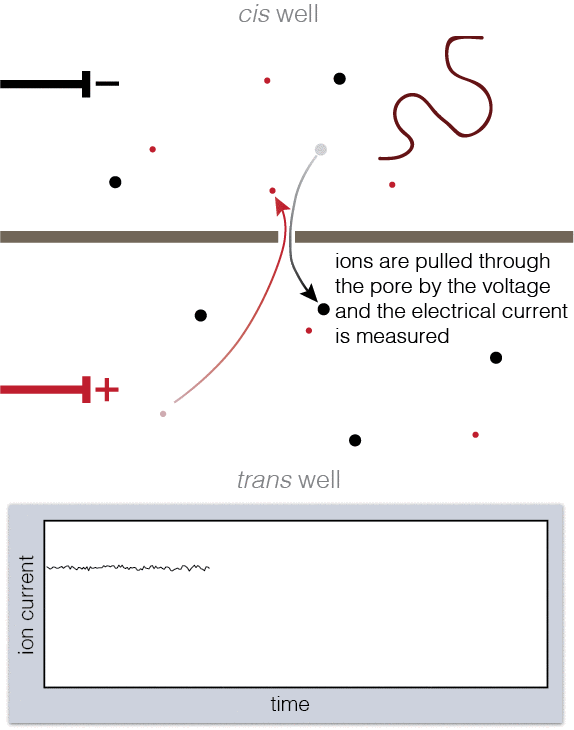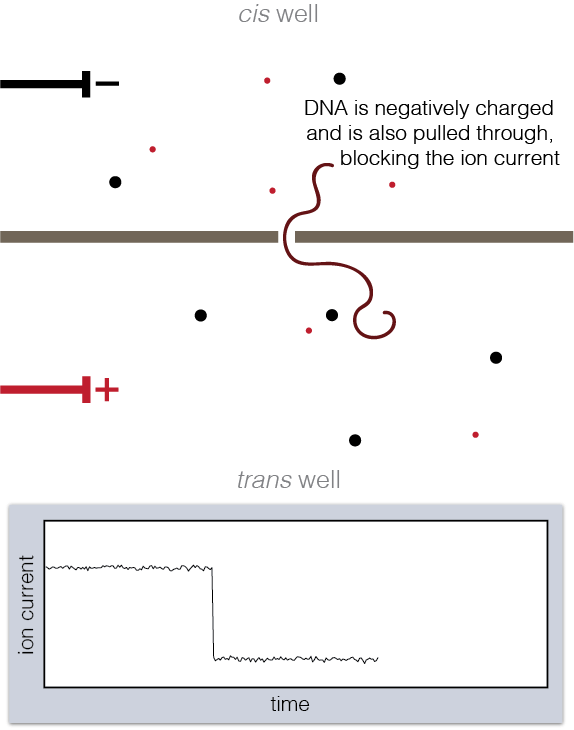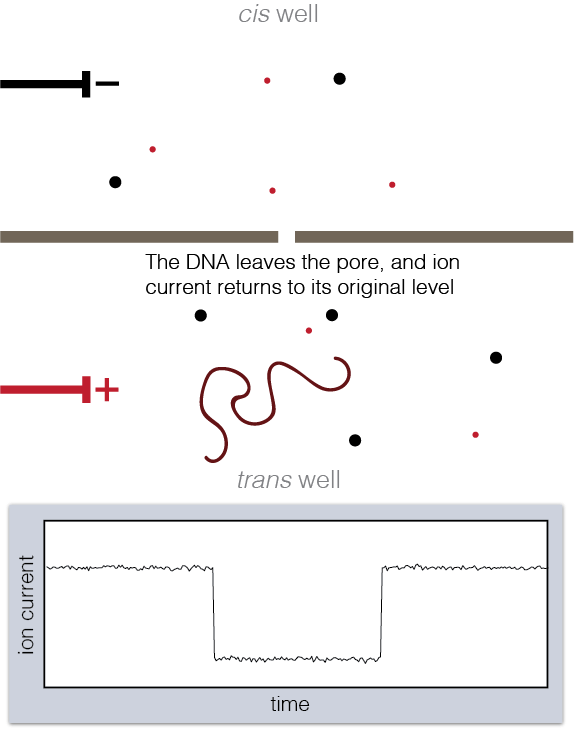
A nanopore is a hole in a membrane on the order of nanometers across. Nanopores are just about the size of biomolecules like DNA or proteins, which makes them perfect tools for trapping and studying the stuff that life is made of.
To set up a nanopore experiment, we put a single nanopore in a membrane separating two wells of a salt solution. Voltage is applied across the membrane, causing salt ions to flow, and this ion current is measured. When large charged molecules like DNA are pulled into the pore, they physically block the ions from flowing through, reducing the observed current. Depending on the DNA bases (A, C, G, or T) blocking the pore, we see a different ion current blockage.
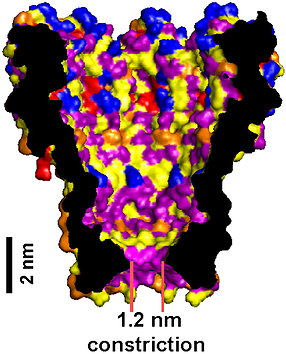 MspA pore. The short and narrow constriction provides exquisite sensitivity to individual DNA bases. Derrington 2010, PNAS.
MspA pore. The short and narrow constriction provides exquisite sensitivity to individual DNA bases. Derrington 2010, PNAS.
Nanopores can be made by punching holes in solid materials like silicon, but these can be hard to make reproducibly. Therefore, we use some existing biological machinery: a biological lipid membrane similar to a cell membrane, together with a membrane protein called MspA, which is normally used by cells to allow chemicals in and out of the cytoplasm.
MspA is an ideal nanopore for studying DNA, because its 1.2 nanometer opening is just large enough for only one piece of single-stranded DNA to thread through at a time, and its short constriction means that only the few DNA bases in that narrowest part are affecting the ion current at a time.
One of our lab's first successes was mutating MspA so that DNA can translocate through it. Natural MspA has negative charges in the constriction, which had to be removed before the also-negatively-charged DNA could pass through.
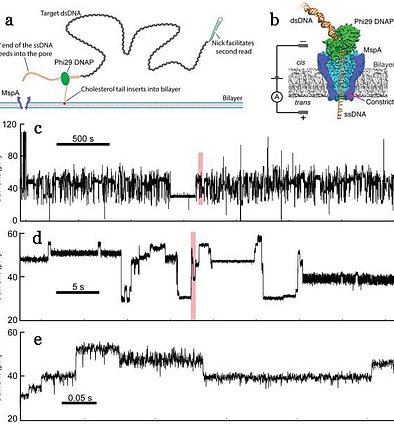 Long nanopore reads of genomic DNA. (a) and (b) An enzyme is used to control translocation, allowing us to observe stepwise motion of DNA through the pore. (c) through (e) This results in steplike current measurements, where each current level corresponds to a new DNA position. Very long reads are possible. Zooming in to each red box on the plot above shows the detail of the nanopore data and the large number of observed DNA steps. Laszlo 2014, Nat. Biotech.
Long nanopore reads of genomic DNA. (a) and (b) An enzyme is used to control translocation, allowing us to observe stepwise motion of DNA through the pore. (c) through (e) This results in steplike current measurements, where each current level corresponds to a new DNA position. Very long reads are possible. Zooming in to each red box on the plot above shows the detail of the nanopore data and the large number of observed DNA steps. Laszlo 2014, Nat. Biotech.
Nanopores are used in a promising next-generation DNA sequencing technology with unique advantages. Since the ion current blockage depends on the DNA bases that are in the pore, we can decode these current signals and sequence DNA.
The DNA ordinarily goes through the pore very quickly, at around a million bases per second. This is too fast to get a read of individual bases, so we have to slow the DNA down. To do this, we use more biological machinery: enzymes that walk along DNA such as helicases and polymerases. These enzymes are too big to go through the pore, so they act like an anchor, holding the DNA and either pulling it up out of the pore or releasing through one base at a time. Using Phi29 DNA polymerase together with MspA led to the first demonstration of single-nucleotide resolution nanopore sequencing in 2012.
 Chemistry of epigenetic markers. Chemical diagram of cytosine, a standard DNA base, and methyl-cytosine, a modified DNA base known to regulate gene transcription. Methyl group highlighted with red circle. Image credit: wikimedia commons.
Chemistry of epigenetic markers. Chemical diagram of cytosine, a standard DNA base, and methyl-cytosine, a modified DNA base known to regulate gene transcription. Methyl group highlighted with red circle. Image credit: wikimedia commons.
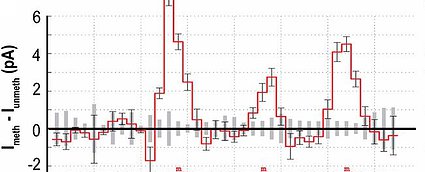 Methylation detection in nanopore reads. Difference between ion current signal from methylated DNA read and unmethylated read. Several current levels around the methylation are strongly affected, allowing for confident mapping of methylation sites; Laszlo 2013, PNAS.
Methylation detection in nanopore reads. Difference between ion current signal from methylated DNA read and unmethylated read. Several current levels around the methylation are strongly affected, allowing for confident mapping of methylation sites; Laszlo 2013, PNAS.
Nanopore sequencing is simple and robust, requiring little in the way of raw materials, expensive equipment, or sample preparation. Nanopore sequencers have even been used on the International Space Station. They are capable of long read lengths, hundreds of thousands of base pairs or possibly more, allowing for sequencing of highly repetitive regions of the genome. Since it is a direct physical measurement rather than a chemical process, nanopore sequencers are readily capable of identifying epigenetic chemical modifications to individual DNA molecules such as methylation and hydroxymethylation, and even sequencing nonstandard DNA bases used in synthetic biology.
Despite all of these advantages, existing nanopore sequencers are challenged by a low single-read sequencing accuracy. Our lab is continuing to work on new solutions that will push this accuracy higher and make nanopore sequencing an everyday tool in medicine and science.
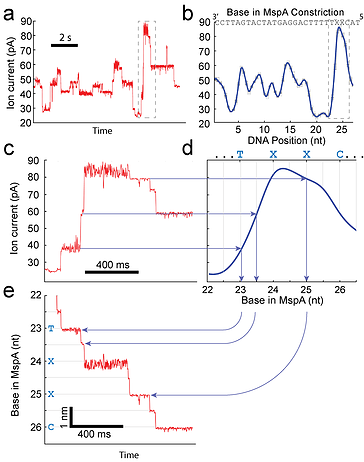 Measuring DNA position with SPRNT. (a) Raw ion current vs. time trace taken with a Hel308 helicase. (b) A consensus of ion currents for the DNA sequence shown in panel a. The dark blue curve is a spline interpolant of the data. This spline acts as a non-linear transformation from ion-current to DNA position. (c) A zoom-in of the dashed box shown in panel a. The horizontal arrows map the raw data to the spline curve, (d). The vertical lines then provide the precise position of the DNA. (e) The fully transformed curve of position vs. time for the trace shown in panel (c). This panel shows that the enzyme Hel308 moves in two sub-nt steps for every nucleotide translocated by the DNA. Craig 2017, PNAS.
Measuring DNA position with SPRNT. (a) Raw ion current vs. time trace taken with a Hel308 helicase. (b) A consensus of ion currents for the DNA sequence shown in panel a. The dark blue curve is a spline interpolant of the data. This spline acts as a non-linear transformation from ion-current to DNA position. (c) A zoom-in of the dashed box shown in panel a. The horizontal arrows map the raw data to the spline curve, (d). The vertical lines then provide the precise position of the DNA. (e) The fully transformed curve of position vs. time for the trace shown in panel (c). This panel shows that the enzyme Hel308 moves in two sub-nt steps for every nucleotide translocated by the DNA. Craig 2017, PNAS.
As a result of our work to understand the underlying physics that makes nanopore DNA sequencing possible, we discovered that we could measure the movement of single enzyme molecules at incredible resolution. We call this technique SPRNT. In SPRNT, a single DNA molecule bound to an enzyme is drawn into MspA.
Once the DNA is captured by the pore, the enzyme controls the motion of the DNA through the pore. As the enzyme moves along the DNA, the motion of the DNA through the pore results in a change in the ion-current passing through the pore because the DNA bases therein affect the ion current.
Even small (sub-nucleotide) movements of the DNA up or down within the pore’s constriction lead to a significant change in the measured ion current. The core concept of SPRNT is that the ion-current through the nanopore provides a precise record of an enzyme’s progression along a DNA molecule (pictured to the right), enabling high-resolution single-molecule measurements of enzymes.
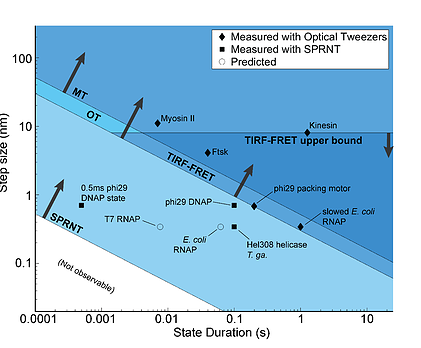 Spatiotemporal resolution of single-molecule techniques. Sensitivity graph of various single-molecule techniques (SPRNT, Optical Tweezers OT, Magnetic Tweezers MT, Total-internal reflection fluorescence FRET TIRF-FRET). The diamonds are enzymes that have been measured with optical tweezers. The squares are enzymes that have been measured with SPRNT; note that these are beyond the resolution of the other single-molecule techniques. The open circles are predictions for enzymes that have not been measured. Laszlo 2016, Methods.
Spatiotemporal resolution of single-molecule techniques. Sensitivity graph of various single-molecule techniques (SPRNT, Optical Tweezers OT, Magnetic Tweezers MT, Total-internal reflection fluorescence FRET TIRF-FRET). The diamonds are enzymes that have been measured with optical tweezers. The squares are enzymes that have been measured with SPRNT; note that these are beyond the resolution of the other single-molecule techniques. The open circles are predictions for enzymes that have not been measured. Laszlo 2016, Methods.
SPRNT can resolve step sizes of less than 0.5 Ångstroms at millisecond time resolution, smaller and faster than other techniques currently in use to measure single enzymes walking along DNA. In fact, many enzymes move so quickly and in such small steps that other single-molecule techniques cannot resolve individual enzyme steps at their physiological speeds (pictured to the left). SPRNT thus enables studies that were not previously possible.
In addition, because the DNA sequence is simultaneously measured using nanopore sequencing, enzyme kinetics behavior can be related to the DNA sequence that is in the enzyme to examine how DNA substrate influences enzyme behavior. SPRNT is high-throughput and parallelizable. A single MspA pore can produce hundreds of single molecule recordings in an afternoon.
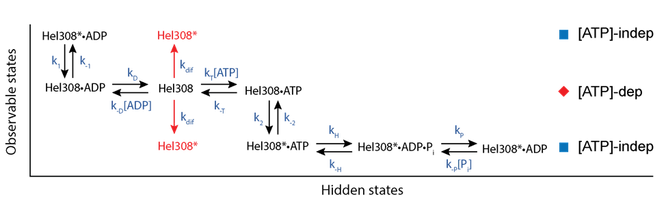 Kinetic Mechanism of Hel308 helicase. In the [ATP]-dependent step (red diamond row) ATP and ADP compete for the Hel308 active site. If ATP binds the enzyme moves forwards along the DNA, whereas if ADP binds the enzyme moves backwards along the DNA. In the [ATP]-independent step (blue square rows), the ATP is hydrolyzed and phosphate is released, before the enzyme proceeds to the [ATP]-dependent with ADP still bound. ADP is then released and the cycle continues. Craig 2017, PNAS.
Kinetic Mechanism of Hel308 helicase. In the [ATP]-dependent step (red diamond row) ATP and ADP compete for the Hel308 active site. If ATP binds the enzyme moves forwards along the DNA, whereas if ADP binds the enzyme moves backwards along the DNA. In the [ATP]-independent step (blue square rows), the ATP is hydrolyzed and phosphate is released, before the enzyme proceeds to the [ATP]-dependent with ADP still bound. ADP is then released and the cycle continues. Craig 2017, PNAS.
Using SPRNT we resolved two distinct kinetic substates of the helicase Hel308, which are not observable by any other technique. We found that one of these states was [ATP]-dependent and the other was [ATP]-independent. Using SPRNT, we uncovered the mechanical and chemical details of how Hel308 uses ATP hydrolysis to achieve directed motion along a DNA substrate, and we placed each of these chemical steps in the context of the two observable Hel308 substates (pictured above).
We are actively working on extending SPRNT to other enzymes, and to date have recorded SPRNT data on 7 different nucleic acid processing enzymes, including helicases, DNA polymerases and RNA polymerases.