Microscopy
Table of Contents
HIV-1 Capsid Assembly
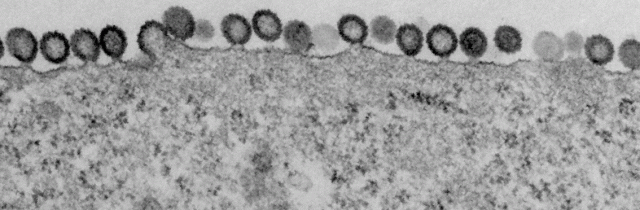
Virus assembling and budding from cells infected with a HIV-1 virus encoding GagZip, a chimeric Gag protein in which the nucleocapsid region of Gag is replaced by a leucine zipper. Lingappa lab & FHCRC EM facility, 2011. For more information, see Klein, Reed, et al. Journal of Virology 85:7419, 2011.
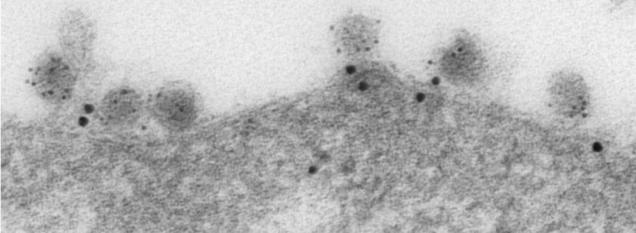
Immunoelectron micrograph showing ABCE1-containing “virus assembly machines” at the plasma membrane of an infected cell. The HIV-1 capsid protein Gag is labeled with small gold (black dots) and the cellular enzyme ABCE1 is labeled with large gold (black dots).

HIV-1 capsids assembling at the plasma membrane. Transmission electron micrograph, The Lingappa Lab & The Fred Hutchinson Cancer Research EM facility, 2006.
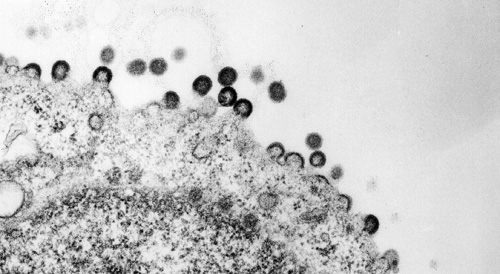
HIV-1 Gag assembling into capsids and budding from the plasma membrane. Transmission electron micrograph, The Lingappa Lab & The Fred Hutchinson Cancer Research EM facility, 2006.
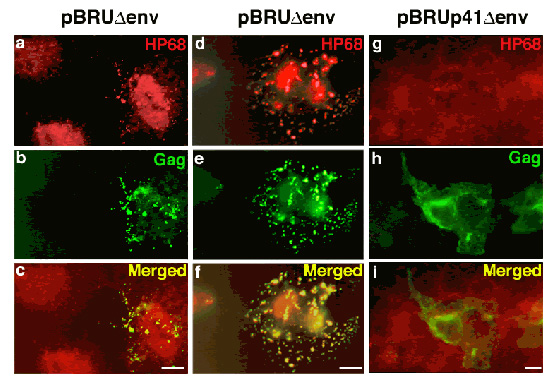
Wild-type Gag, but not an assembly-defective Gag mutant, recruits the cellular protein ABCE1 to sites of assembly:
Cos-1 cells were transfected with a plasmid encoding the full-length, wild-type HIV-1 pBru genome with a deletion in Env (first two columns, a – f), or a pBru plasmid encoding a truncated form of HIV-1 Gag termed p41 that fails to associate with ABCE1 or assemble into capsids (third column, g – I). Immunostaining was performed for HIV-1 Gag (green; Cy-2) and endogenous ABCE1 (red; Cy-3). Each column shows one field of cells examined by immunofluorescence microscopy using filters that allow visualization of either ABCE1 (a, d, g) or Gag (b, e, h) or both (c, f, I). The first column shows two cells on the left that are not expressing Gag (b). In these cells, endogenous cellular ABCE1 is present diffusely (a). In contrast, the cell on the far right in the first column (b) and the cell in the center of the middle column (e) both express Gag in a coarsely punctate pattern than most likely represent sites of Gag assembly at the plasma membrane. In both cases endogenous ABCE1 takes on a coarse punctate appearance (a, d). Merged images reveal that ABCE1 is co-localized with Gag in these cells (c, f). In cells with low levels of Gag expression, a large pool of diffuse ABCE1 is still present in addition to a small pool of punctate ABCE1 (cell on right in a, c, e). In cells expressing higher levels of Gag, a larger percentage of endogenous ABCE1 has been recruited into punctate sites of assembly, and the residual pool of diffuse ABCE1 is much smaller. As expected, the p41 Gag mutant fails to form punctate sites of assembly (h) or to recruit HP68 from diffuse pools (g) as seen by failure of co-localization (i). (From Zimmerman et al., Nature 415(6867), 88-92 Fig. )
HCV Capsid Assembly
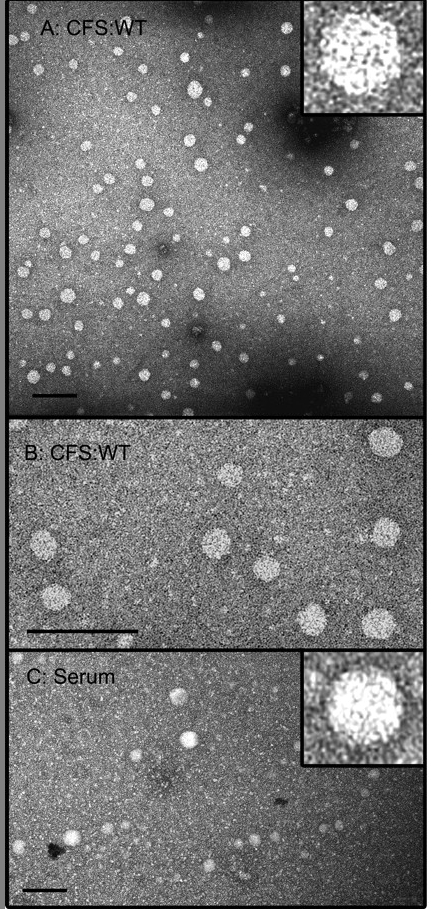
The 100S HCV capsids from the cell-free system (CFS) are morphologically similar to authentic capsids. (A and B) Cell-free reactions programmed with C191 transcript were separated by velocity sedimentation, and fractions 6, 7, and 8 (containing 100S capsids) were pooled, dialyzed against phosphate-buffered saline, and subjected to negative-stain TEM. (C) Serum from an infected patient, treated with detergent to remove envelopes, was processed in parallel. Bars, 100 nm. From Klein, Polyak, and Lingappa, J. Virol. 78:957 (2004).
HBV Capsid Assembly
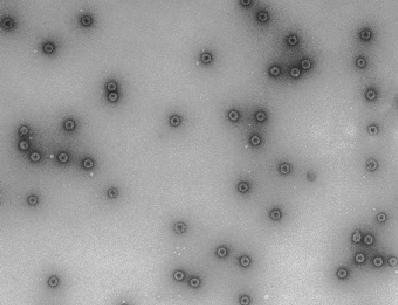
HBV capsids assemble in the cell-free system via a single high molecular weight assembly intermediate:
In a wheat germ cell-free system Hepatitis B virus (HBV) core protein assembles into paricles that are indistinguishable from authentic HBV capsids by biochemical and morphologic criteria (fig. 8). Dissection of this cell-free system reveals that HBV capsids assemble via a single large assembly intermediate that can be isolated and “chased” to form completed capsids. Cellular factors that have yet to be defined are present in this high molecular weight complex.