Research Background
Table of Contents
Overview of HIV Assembly
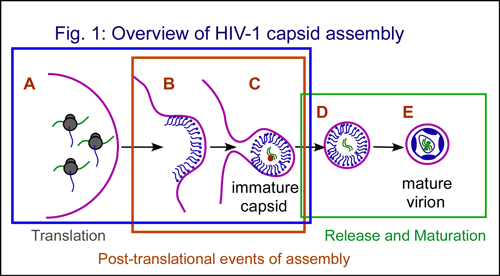
Viruses use host cells to produce new viruses that can infect other cells. A newly released virus contains the viral genome surrounded by a protective capsid shell. In the case of HIV, a single virus contains two copies of the HIV-1 genomic RNA surrounded by more than 3000 copies of the HIV Gag protein. Gag proteins are synthesized (translated) in the cytoplasm (Fig. 1A) and subsequently target to the plasma membrane (Fig. 1B), where they assemble into spherical immature capsids (Fig. 1C). During assembly, Gag must also find and package (encapsidate) the viral genomic RNA. The immature capsid subsequently releases from the cell by budding from the plasma membrane (Fig. 1D). In the last step of virus formation, Gag proteins in the immature capsid are cleaved by the HIV protease, resulting in formation of the mature virus particle (Fig. 1E).
Our Central Hypothesis: Virus Assembly is Catalyzed by Host Enzymes
Our central hypothesis is that viruses co-opt host machinery and use energy (ATP) to facilitate capsid assembly. While capsid proteins can “self assemble” when they are at high concentrations under very specific conditions in test tubes, such spontaneous assembly events are unlikely to occur in the cytoplasm where there are many barriers to self assembly including the low concentration of capsid proteins and the high concentration of cellular proteins. Thus, it is likely that in cells, viruses hijack cellular machinery during capsid assembly to ensure efficient virus production.
Evidence that HIV-1 co-opts cellular machinery to facilitate assembly of the immature capsid came from our finding that ABCE1, an ATP-utilizing cellular enzyme, facilitates HIV-1 capsid assembly (Zimmerman C et al., Nature 415:88, 2002). Our studies also suggest that other primate lentiviruses, such as HIV-2 and SIV, utilize ABCE1 (Dooher J et al., J Virol 78:1645, 2004). More recently, we used siRNA knockdown and rescue approaches to show that another cellular enzyme, DDX6, also facilitates HIV-1 capsid assembly in infected cells (Reed J et al., J Cell Biol 198:439, 2012).
HIV Hijacks Host RNA Granules to form “Virus Assembly Machines”
The cellular machinery that is co-opted by HIV-1 belongs to a class of cellular complexes called “RNA granules” (Reed J et al., J Cell Biol 198:439, 2012). HIV-1 hijacks these cellular RNA granules, transforming them from machines that work for the host into “virus assembly machines”, which are also called assembly intermediates. When Gag co-opts these RNA granules, it forms a pathway of assembly intermediates, each representing a different stage in the assembly of an immature HIV-1 capsid (Fig. 2). The sequential assembly intermediates are named by their S value, which indicates their size (~10S, ~80S, ~150S, ~500S); together with the completed immature capsid (which is ~750S), they form the HIV-1 capsid assembly pathway (Fig. 2). When the immature capsid is completed, it lets go of the cellular machine (RNA granule), which stays inside the cell while the assembled capsid undergoes budding and is released to the outside of the cell.
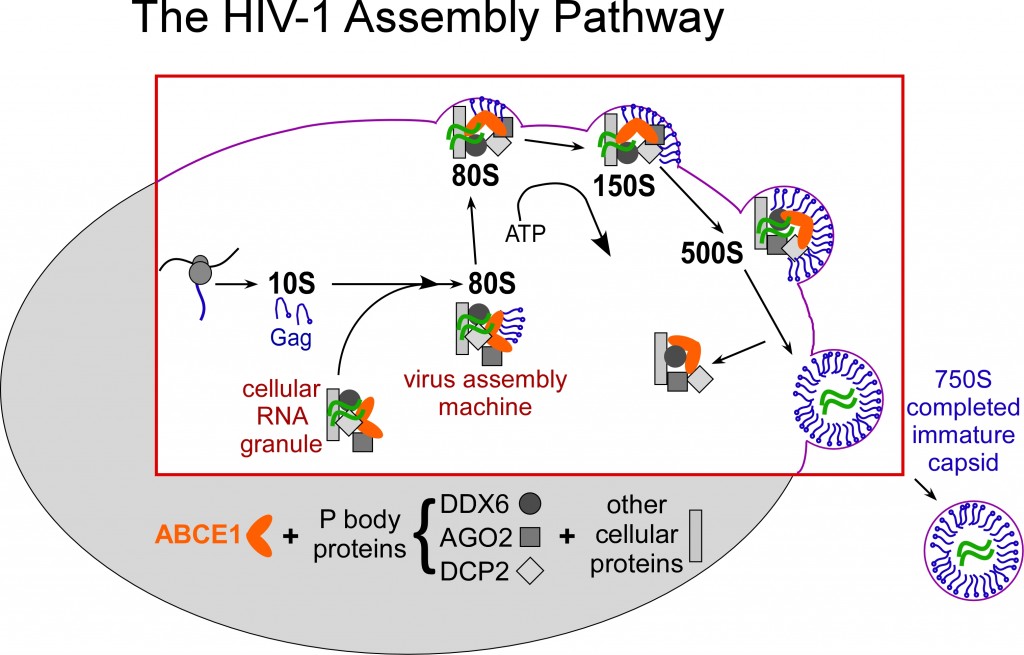
HIV-1 Gag (in blue) hijacks the RNA granule “cellular machine”, transforming it into a “virus assembly machine” (assembly intermediate). The assembly intermediate grows in size (from ~80S to ~150S to ~500S) as the immature capsid is assembled.
In HIV-expressing cells, the different Gag-containing assembly intermediates are formed sequentially over time, as observed in pulse chase experiments and confirmed using immunoelectron microscopy (Dooher et al., Traffic 8:195, 2007).
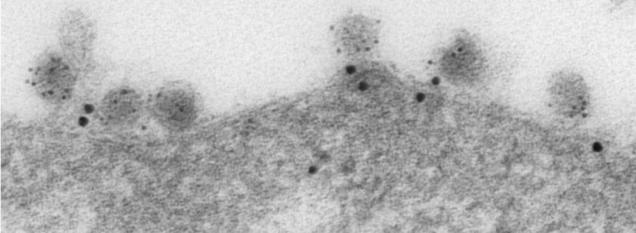
Immunoelectron micrograph showing ABCE1-containing “virus assembly machines” at the plasma membrane of an infected cell. The HIV-1 capsid protein Gag is labeled with small gold (black dots) and the cellular enzyme ABCE1 is labeled with large gold (black dots).
Immunoelectron micrograph showing ABCE1-containing “virus assembly machines” at the plasma membrane of an infected cell. The HIV-1 capsid protein Gag is labeled with small gold (black dots) and the cellular enzyme ABCE1 is labeled with large gold (black dots).
Studies of Gag mutants that are arrested at different steps in the assembly pathway show that HIV-1 assembly intermediates are initially formed in the cytoplasm of HIV-1 expressing cells and then traffic to the plasma membrane, where assembly is completed and budding is initiated (Robinson B et al., J Virol 88:5718, 2014; Fig. 2).
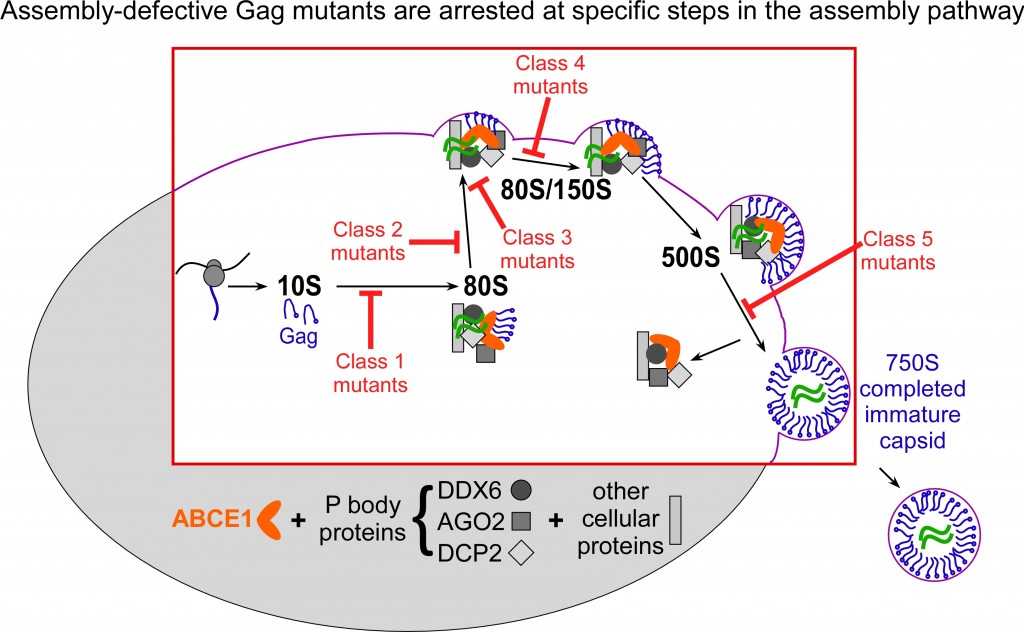
When assembly-defective Gag mutants co-opt cellular machines, they get stuck at specific points in the assembly pathway (shown in red) and are unable to effectively use the cellular machines to complete immature capsid assembly.
Next steps in Understanding “Virus Assembly Machines”
Current studies in the Lingappa lab are addressing how cellular proteins in the “virus assembly machines” (assembly intermediates) facilitate HIV-1 assembly and packaging of the HIV-1 genome. These studies will shed light on why HIV-1 hijacks the poorly understood ABCE1-containing cellular machines (RNA granules) present in the host, and how viral exploitation of these machines impacts HIV-1 pathogenesis in infected individuals.
Why study host machinery involved in virus assembly?
Finding and understanding the cellular machinery co-opted by viruses during assembly could lead to identification of targets for future antiviral drugs. Evidence that these “virus assembly machines” are excellent drug targets comes from the recent identification of potent drug-like compounds that target ABCE1-containing complexes and inhibit rabies virus infection in cells (Lingappa, U et al. PNAS 110:E861, 2013).
Additionally, we need to better understand how these cellular machines function in normal, uninfected cells. The cellular machines (RNA granules) co-opted by HIV-1 are different from other RNA granules (Reed J et al., J Cell Biol 198:439, 2012), in that they are smaller than well-studied RNA granules (e.g. P bodies and stress granules) and contain ABCE1, which has not been found in the better-understood RNA granules. Studies suggest that malfunction of these cellular machines can lead to chronic diseases. Drugs that target these “aberrant cellular machines” are being developed by Prosetta Biosciences.