research

Our recent work has drawn from molecular genetics, genomics, physiology and synthetic biology to build new tools to study signaling dynamics and to apply these tools to a variety of fundamental questions in cell and developmental biology. Specifically, we are:
- Building tools to study and reprogram signaling dynamics
- Integrating metabolic status into growth control networks
- Evaluating the impact of evolution on signaling networks
1. Building tools to study and reprogram signaling dynamics
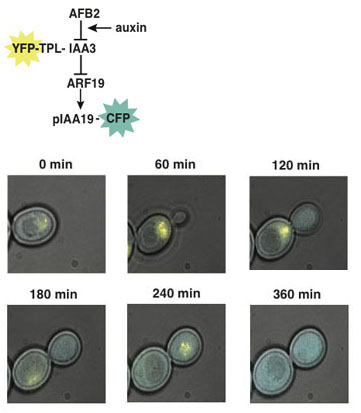
Building dynamic networks from the ground up. Auxin is a plant hormone that plays a key role in nearly every aspect of plant biology. Direct experimental tests of signaling dynamics in this crucial pathway are confounded by the ubiquity of auxin response in plant cells. In collaboration with Eric Klavins in the UW Electrical Engineering Department, we have developed an alternative approach where we are systematically transplanting the auxin response pathway from Arabidopsis into the single-celled yeast Saccharomyces cerevisiae. An analogy to our approach is trying to understand how a radio works by removing components one by one, reconnecting each part in a simple setting, and characterizing the resulting circuits in great detail. We have successfully transferred the nuclear auxin response pathway from auxin perception through activation of transcription—a rather remarkable feat highlighting the fundamental conservation of core eukaryotic cell biology. We are currently excited to apply this system to fundamental control points of signaling, including protein degradation, transcriptional repression and transcriptional activation. We are also developing and deploying new tools to reparameterize core hormone-regulated networks (including auxin, jasmonates and gibberellins) using synthetic transcription factors.
2. Integrating metabolic status into growth control networks
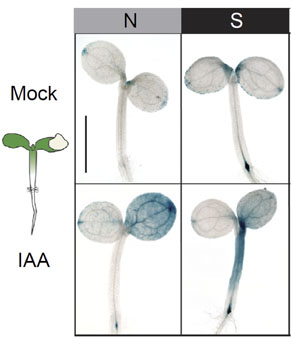
We have discovered that many facets of growth are exquisitely sensitive to developmental stage, as well as genetic and environmental perturbations. Among our most surprising findings was that carbon availability had a dramatic effect on multiple aspects of growth dynamics. Excitingly, the light-regulated transcription factors PIF4 and PIF5 were required for the growth promoting effects of elevated CO2, and the PIFs acted at least in part by regulating the amount of auxin delivered from shoots to the roots. Our results point to direct integration of the light signal downstream of both the phytochrome photoreceptors and photosynthesis. This work provides an outstanding opportunity to integrate cell signaling into an organismal framework of plant growth control. Our progress has been greatly accelerated by an on-going collaboration with Soo-Hyung Kim in UW School of Environmental and Forest Sciences.
3. Evaluating the impact of evolution on signaling networks
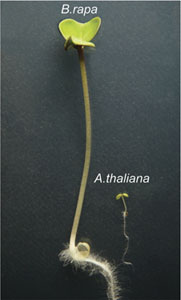
Approaches for engineering new crop varieties are remarkably crude compared to the design and implementation of non-biological technology. One strength of engineering is its ability to parse complex systems, such as a Boeing 787, into sub-networks or modules that can be analyzed in isolation. The synthetic auxin response system in yeast, developed by my lab in collaboration with Eric Klavins in the UW Electrical Engineering Department, makes it possible to interrogate the function of plant auxin signaling modules in isolation. We are currently testing the function of auxin components of Zea mays and Brassica rapa in our synthetic system. This comparative approach may help answer one of the oldest questions in auxin biology: how does such a simple molecule do so many different things?























































































