Many sites of the human body are colonized by complex communities of microbes (the "human microbiome") in both health and various disease states. Highly diverse, polymicrobial specimens are often difficult, or even impossible, to fully characterize by techniques in common clinical use:
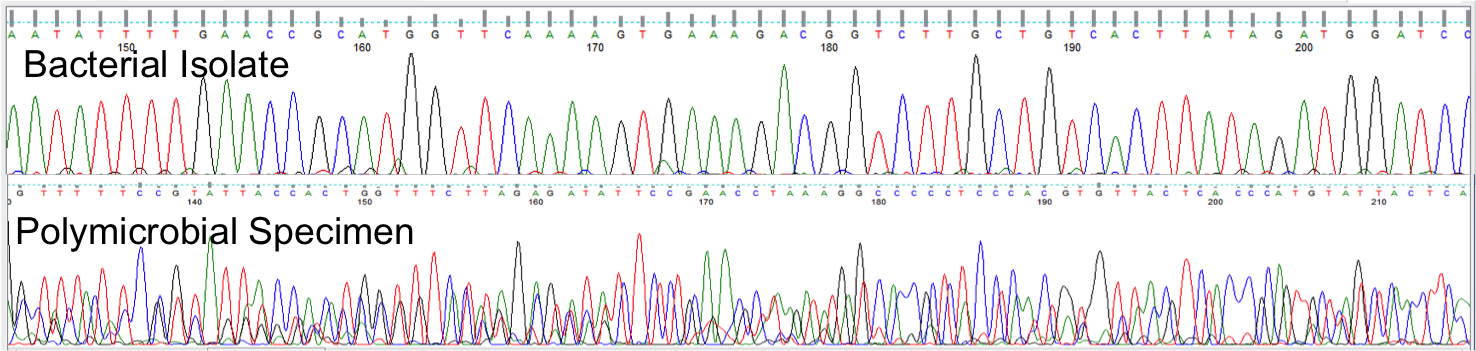
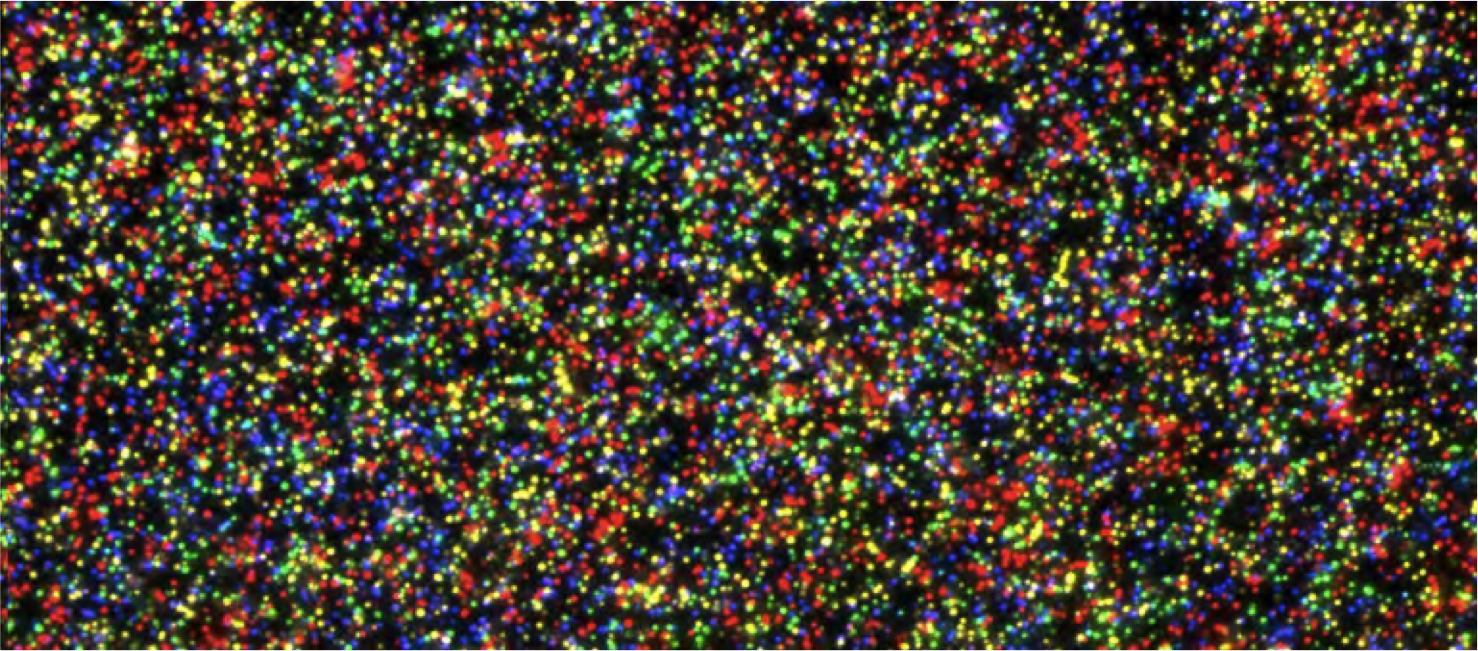
In contrast to conventional approaches, next-generation DNA sequencing (alternatively termed "NGS", "high-throughput sequencing", "massively parallel sequencing", or "deep sequencing") provides independent sequence data from millions of individual DNA molecules (Figure 2), allowing each fragment to be classified independently.
This unique ability extends upon the advantages of current molecular methods by allowing us to catalog the organisms present within even very complex polymicrobial bacterial communities, directly from patient specimens.
Our lab currently offers high-fidelity Illumina next-generation DNA sequencing of clinical specimens which contain multiple bacterial DNA templates. Methods are validated for the purpose of clinical molecular diagnosis and patient care. Research services are also available - please contact us for additional information.
This test is available as reflex testing for specimens which are expected to be polymicrobial based on broad range bacterial PCR.
To learn more about clinical applications and methodology, please see the following publications from our lab:
Contact molmicdx@u.washington.edu for details or questions.
Upon completion of testing, a report is issued describing the results of 16S next generation sequencing. To view a sample report, click here or the thumbnail at left.
For additional information on how to submit a request and recieve a report, please contact us!