
How to make 6000 GAL4-AD clones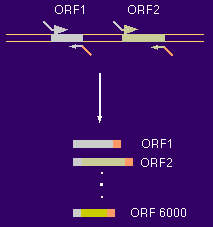 For each of the ~6000 yeast ORFs a specific primer pair was synthesized (Research Genetics, Alabama) and the ORF amplified from genomic DNA. Each of the forward primers had a specific common tail as much as the reverse primers, which had a different common tail (20 nucleotides). 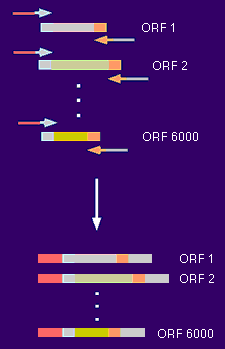 The common tails served as priming sites for a second round of PCR (the "rePCR"). The primers for this re-PCR were 70mers with 50 nucleotides of yet another set of common tails. Since all primary PCR products had already common 20mers at their ends, one pair of 70mers was sufficient to re-PCR all 6000 ORFs. 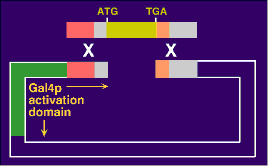 The re-PCR products with the common 50 mers were then used to recombine the ORFs into two-hybrid vectors in frame with the GAL4 activation domain. Recombination cloning can be achieved by simply transforming linearized vector and PCR product into yeast. The only requirement for recombination is an overlap between insert and vector of at least 30-40 base pairs. Return to Protocols Page |
|
HHMI, |