Engineering RNA-binding proteins for biology
RNA-binding proteins often have modular structure: the RNA-binding activity is contained within well-identifiable structural domains that target the RNA to be processed, exported or localized. The most common of these domains is the RRMs, which is present in hundreds of copies in all eukaryotic genomes. It has so far been impossible to engineer RRMs to target non cognate sequences, as is now routinely done for zinc fingers, but we have recently been able to do exactly that. We are using protein design and engineering tools we have developed to redirect RRMs to target different microRNA precursors and study their biogenesis. Once the activation and repression domain within Fox protein isoforms are identified, we will have a modular set of tools (RNA-binding box, activation box and repression box) that can be mixed and matched in different proteins to affect and regulate production of microRNAs exogenously.
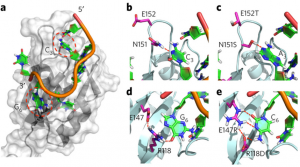
Engineering the RNA-binding specificity of Rbfox-RRM by rational design. (Chen, Y., et al. Nat Chem Bio 2016)
The structural molecular biology of microRNA processing
MicroRNAs are short non coding transcripts that regulate protein production by binding to mRNAs and causing increased RNA turnover or inhibition of protein synthesis. There are >1,000 microRNA in humans which regulate >70% of the transcriptome. MicroRNAs are produced as long precursors that are processed first in the nucleus and then the cytoplasm by two RNAse III enzymes, Drosha and Dicer. Processing is regulated by unclear structural and dynamic aspects of the precursor species, and by the interaction of the precursor RNAs with RNA-binding proteins. We have identified new regulatory activities of pre-mRNA splicing factors that interact with and regulate processing of certain microRNAs in a gene-specific matter, and we now seek to establish the structural and biochemical basis for regulation and examine the physiological consequences of this regulation.
Namely, we have found that the Fox family of alternative splicing factors (Rb-Fox) regulate the expression of microRNAs containing Fox binding sites in vitro and in vivo and have established the structural basis for this function. Intriguingly, different isoforms of Fox protein can up or down regulate production of the same microRNAs, suggesting that the RNA-binding activity can be functionally and probably biochemically separated from activation or inhibition of microRNA processing. What are the domains responsible for regulation? Are they transferrable to different RNA-binding proteins? What is the mechanism of regulation? What other factors are involved?
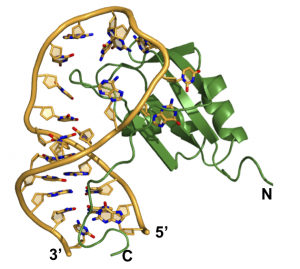
Most importantly, what are the biological consequences of these interactions? Mis-regulation of Fox-1 is observed in the brain of autistic patients; does the effect on the brain arise from misregulation of maturation of a key set of microRNAs? The RNA-binding sequences for other RNA-binding proteins are conserved within pre-miRNA precursors, including miRNAs expressed in cancer (onco-miRs). Does the transforming activity associated with these other microRNAs arise from mis-regulation of their maturation by RNA-binding proteins?
Structural basis for recognition of pre-miR-20b by the Rbfox RRM (Chen, Y., et al. NAR 2016)
Inhibition of microRNA processing in chronic disease
Many microRNAs are over-expressed in diseases such as cancer (e.g. miR-20b or miR-21) or regulate essential metabolic activities, for example fatty acids in the blood circulation (miR-33) or insulin production (miR-103 and miR-107). Therapeutic targeting of certain mature microRNAs (e.g. miR-21 and miR-107) with oligonucleotide analogues has been shown to revert the disease phenotype in cells and sometimes primate models. Thus, inhibiting the maturation of specific microRNAs to reduce the level of mature RNAs is likely to be therapeutically beneficial, especially if we could use chemistry other than oligonucleotides, to avoid the well-known limits of that chemistry in delivery, pharmacology and cost.
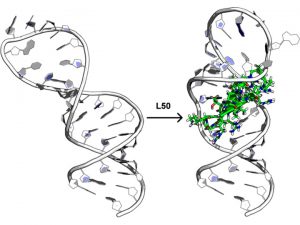
A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing (Shortridge, M., et al. ACS Chem Biol 2017)
We have identified a class of proteolytically stable, cell permeable cyclic peptides that bind to human pre-miR-21 by targeting the terminal stem-loop region of the primary precursor species. These peptides reduce microRNA processing in vitro and in cells by modulating regulatory RNA-protein interactions, we hypothesize. By using structure based approaches, we want to optimize the activity of these initial hits to inhibit pre-miR-21 processing and reduce miR-21 activity in cancer cells, exactly as we have done, using the same chemistry, in another project targeting the HIV RNA regulatory element TAR (see below). Success in this project would open up a new approach to inhibiting miRNA with broad biological implications and considerable potential for commercialization.
Inhibition of viral replication
The ability of HIV to enter a latent state creates a fundamental obstacle to the eradication of infection by anti-retroviral therapy and has prompted continued interest in developing inhibitors that block the re-emergence of latent proviruses. Such an inhibitor would be able to block viral replication both in acutely and in chronically infected cells and ideally target the reservoir of slowly replicating viruses that persists in the presence of Highly Active Anti Retroviral Therapy (HAART).
We have identified cyclic peptides mimis of the transactivatpr protein Tat that bind to the HIV TAR RNA (Fig. 2) with unprecedented low picomolar activity by expanding peptidic chemistry to include un-natural side chains. The lead peptidomimetic compounds penetrate cellular membranes readily, have no cytotoxicity and inhibit viral replication in primary lymphocytes with activity slightly better than AZT. We are optimizing the cellular activity of the cyclic peptides further to inhibit the function of TAR by exploiting un-natural peptide side chains and modifications of the peptide backbone; by establishing the molecular mechanism of action of the peptide leads; by using advanced structure-based design methods to turn the peptides into small molecules armed with the knowledge provided by the structures of the peptides of the critical point of intermolecular contacts.
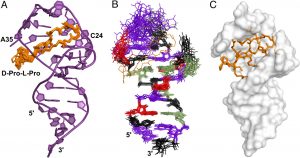
Structure of peptidomimetics inhibitors of HIV replication bound to HIV-1 TAR RNA (Davidson A., et. al., PNAS 2009)
The structural biology of mRNA 3’-end processing
RNA processing reactions can be reconstituted in vitro, but they occur more efficiently in the cell because they are closely integrated with transcription. How these biological processes are regulated and integrated with each other remains unclear at the molecular level and poorly understood structurally. We study the molecular basis for the specific recognition of different phosphorylatyed forms of the C-terminal domain of RNA polymerase II, and how a new phosphatase generates the form of the CTD found at the 3’-end of genes. We study the molecular basis for RNA recognition by RNA processing factors as well, but the biggest prize we are pursuing, using a combination of experimental approaches (NMR, x-ray, SAXS and EM), is the structure of the megadalton size complex of proteins responsible to specify the processing site and execute cleavage and polyadenylation (below).
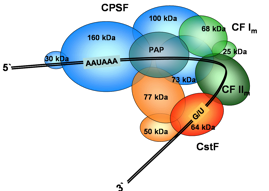
Vertebrate 3’-end processing complex; the structure of many components is now known, but how they are assembled and regulated remains unknown
Structure of non-coding RNAs
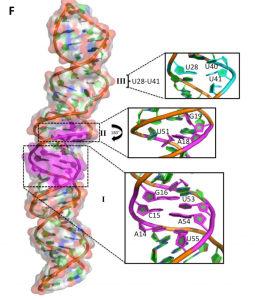
We investigate the structure of long non-coding RNAs, widely expressed and numerous RNAs with uncertain function and unknown structures. Specifically, we are investigating the structure of an exceptionally well-conserved long non coding RNA that play critical roles in Zebrafish development (we have identified a conserved secondary structure that we are characterizing biologically and structurally); of an RNA thermometer from N. mengingitis that allows immune evasion by controlling expression of capsule genes over a narrow temperature range (we have established the mechanism of action of the thermometer and are establishing its structure); and of promoter associated RNAs that regulate transcription of proto-oncogenes.
Structural analysis of the CssA thermometer. (Barnwal, R., et. al. NAR 2016)
We approach this complex structural problem by using chemical and enzymatic methods to probe the RNA secondary structure that we confirm using sequence variation and co-variation. We define conserved structural elements to validate by sequence analysis and biologically through our collaborations. We use lower resolution structural techniques such as EM and SAXS to define the envelope of the structure, and higher resolution approaches (NMR and crystallography) to establish the structure of conserved and functionally important domains. By combining methods that investigate RNA structure at multiple scales and level of resolution and integrating them with biological experiments, we plan to map the structure-function relationships for these and, in the future, other important lncRNAs.
Perspectives
The structural biology projects illustrated above are and will remain a central part of our future research interests. I anticipate that the projects on long non coding RNAs will grow in importance in the near future: there is so much to discover about the structure/function relationship of these RNAs and so many lncRNAs to study. In turn, this knowledge would open up new opportunities to target these RNAs in disease conditions. However, if we were truly able to develop the chemistry of RNA targeting with peptides and small molecules, by reaching beyond cell based assays and into pre-clinical and clinical evaluation, the implications would be very broad and genuinely ground breaking.