Welcome to the Chiu Research Group!
My lab is focused on developing new methods for probing complex biological processes at the single-cell and single-molecule level, and on applying these new techniques for addressing pressing biological problems. Listed below are some examples.
|
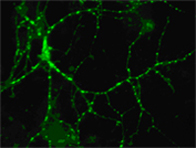
We are developing high-resolution and quantitative microscopy tools to study cellular systems at a higher level of precision than is possible with conventional techniques. For example, fluorescence-microscopy images often contain puncta in which the fluorescent molecules are spatially clustered. We have developed a statistical framework that uses single-molecule intensity distributions to deconvolve the number of fluorophores present in fluorescent puncta as a way to “count” protein numbers present. This method has the advantages of single-molecule measurements, providing both the mean and variation in molecules per puncta (see e.g. ref 78 and 126 on publication list). The image to the left shows the punctate appearance of the fluorescence emission from a pyramidal neuron. |
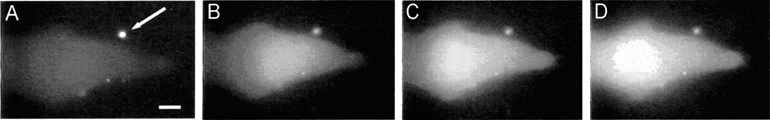
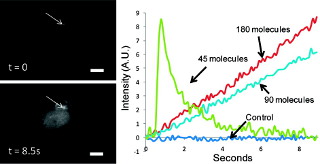
We are researching new techniques that use optics and nanomaterials for the spatiotemporally resolved analysis and perturbations of cells. The sequence of images to the right shows one example, where a single fluorescently labeled lysosome was surgically removed from a B cell using the polarization-shaped optical vortex traps that we developed (see e.g. ref 76). Below is another example, where we integrated optical control with nanocapsules (arrow) to achieve highly localized delivery of bioactive molecules to a cell. (A) shows the image just prior to delivery of a biological stimulus; (B-D) shows the increase in intracellular calcium, visualized as a fluorescence increase, across the cell after the stimulus has been released from the nanocapsule (see e.g. ref 36).
|  |
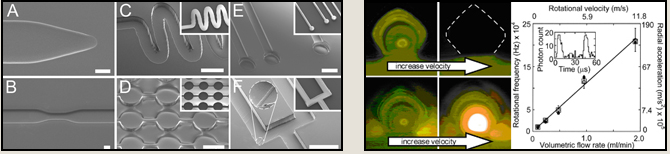
We are active in developing new techniques in microfabrication and microfluidics so we can interface effectively with single cells and control cellular behavior. We have developed, for example, two new materials that we termed TPE (thermoset polyester) and PUMA (polyurethane-methacrylate) for the rapid prototyping of microdevices (see images A-F at bottom left) (see e.g. ref 54, 58, 101, 102). Another example is the finding that ultrahigh radial accelerations (> million gs) can be generated in microvortices (see two panels to the bottom right) (see e.g. ref 43). |
 |
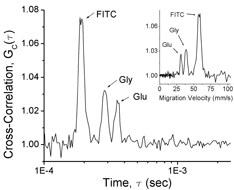
| We are currently engaged in developing a new technological platform by which we can extract detailed chemical information from subcellular structures and organelles, while preserving the spatiotemporal information offered by high-resolution microscopy. One component of this technology relies on the ability to control the reactivity of a single or a small number of molecules as well as to separate and identify the individual molecules. To accomplish this, we are employing femtoliter-volume aqueous droplets that can be generated on-demand using fluidic techniques (see e.g. ref 59, 100). The images to the left show a single cell and a single mitochondrion, respectively, confined to an aqueous droplet. The plot to the right shows the result of single-molecule capillary electrophoresis in which the retention time was measured molecule by molecule (see e.g. ref 77). |
|
|
|
|


 We are engaged in a wide range of nanomaterials design and synthesis, and assembling these nanomaterials into controlled architectures with the desired functions. One example is the light-addressable nanocapsules that we have developed over the past years for intracellular delivery of bioactive molecules (see e.g. ref 124). The panels to the left
We are engaged in a wide range of nanomaterials design and synthesis, and assembling these nanomaterials into controlled architectures with the desired functions. One example is the light-addressable nanocapsules that we have developed over the past years for intracellular delivery of bioactive molecules (see e.g. ref 124). The panels to the left 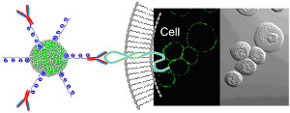 show the intracellular release of IP3. Another example is the highly fluorescent polymer nanoparticles that we are designing and developing for cellular studies. The panels to the right show the cellular targeting using bioconjugates based on these new polymer nanoparticles (see e.g. ref 121, 122).
show the intracellular release of IP3. Another example is the highly fluorescent polymer nanoparticles that we are designing and developing for cellular studies. The panels to the right show the cellular targeting using bioconjugates based on these new polymer nanoparticles (see e.g. ref 121, 122). We are applying our new techniques, in combination with established molecular biology methods, to address two important biological areas: (1) Synaptic transmission and neuronal function, and (2) Single-cell cancer biology. The fluorescence (left panel) and AFM (right panel) images to left show individual synaptic vesicles that we have isolated from mouse brain for detailed single-molecule studies. The images to the right show a single cancer cell that we have isolated from a patient sample.
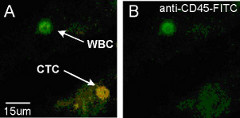
We are applying our new techniques, in combination with established molecular biology methods, to address two important biological areas: (1) Synaptic transmission and neuronal function, and (2) Single-cell cancer biology. The fluorescence (left panel) and AFM (right panel) images to left show individual synaptic vesicles that we have isolated from mouse brain for detailed single-molecule studies. The images to the right show a single cancer cell that we have isolated from a patient sample.