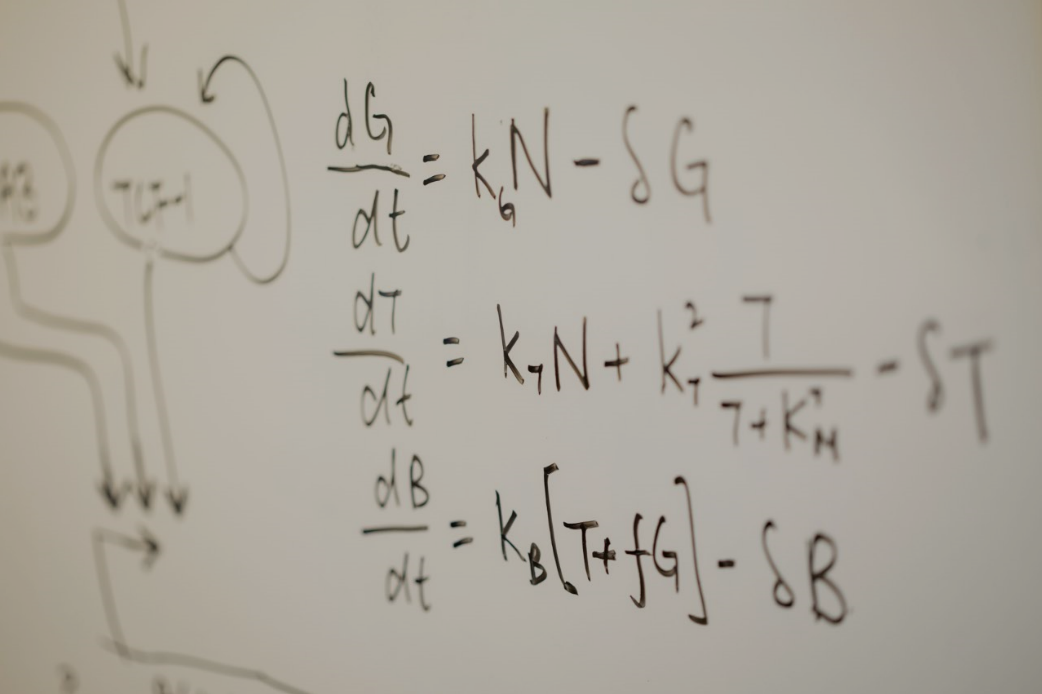Selected Preprints

A timed epigenetic switch balances T and ILC lineage proportions in the thymus
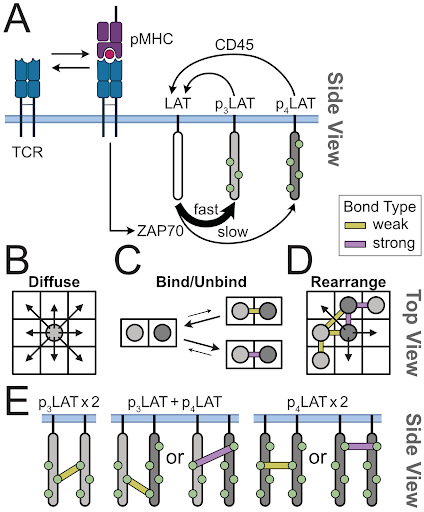
Proofreading and single-molecule sensitivity in T-cell receptor signaling by condensate nucleation
Selected Publications
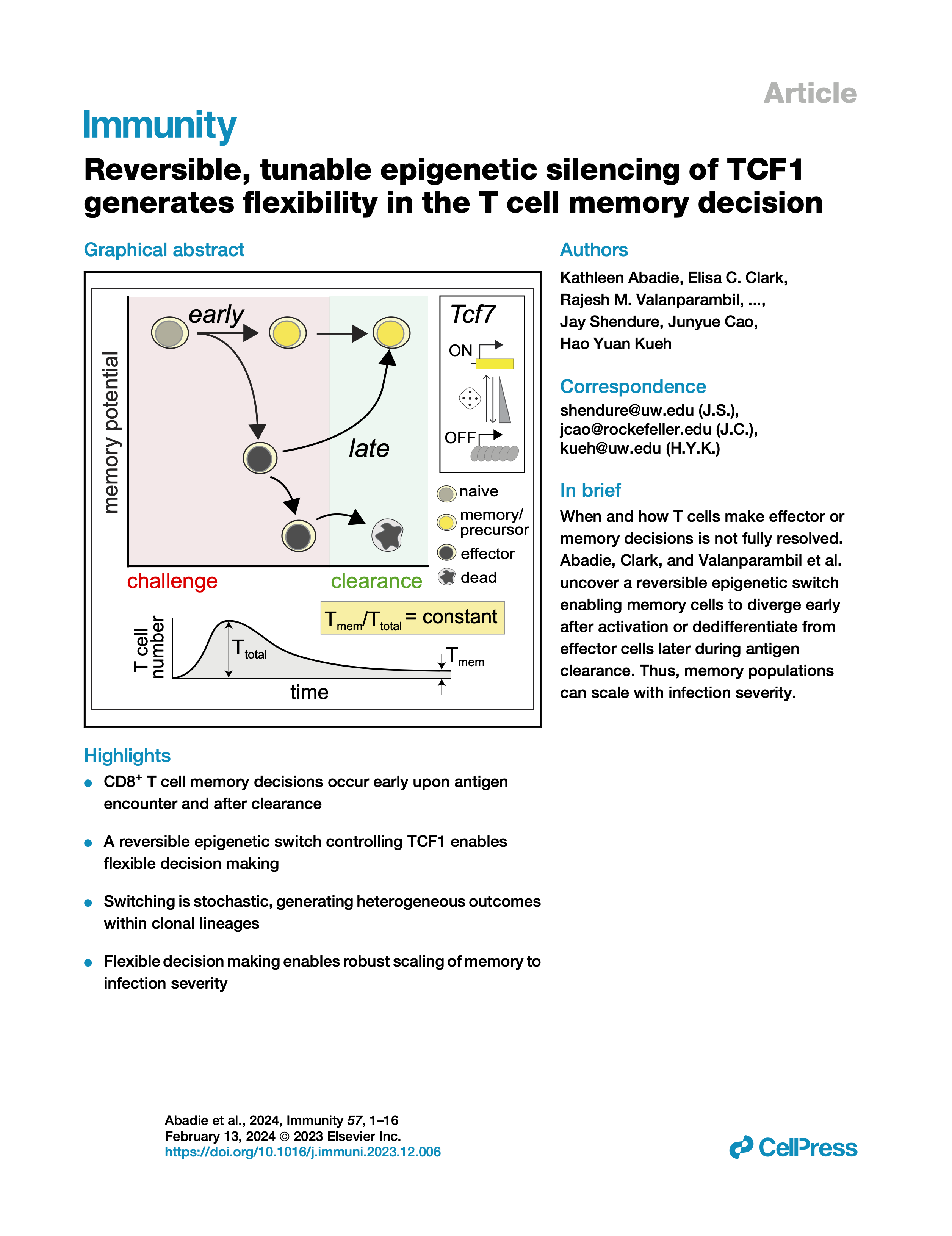
Reversible, tunable epigenetic silencing of TCF1 generates flexibility in the T cell memory decision

Antigen perception in T cells by long-term Erk and NFAT signaling dynamics

Unsupervised discovery of dynamic cell phenotypic states from transmitted light movies

Scalable control of developmental timetables by epigenetic switching networks
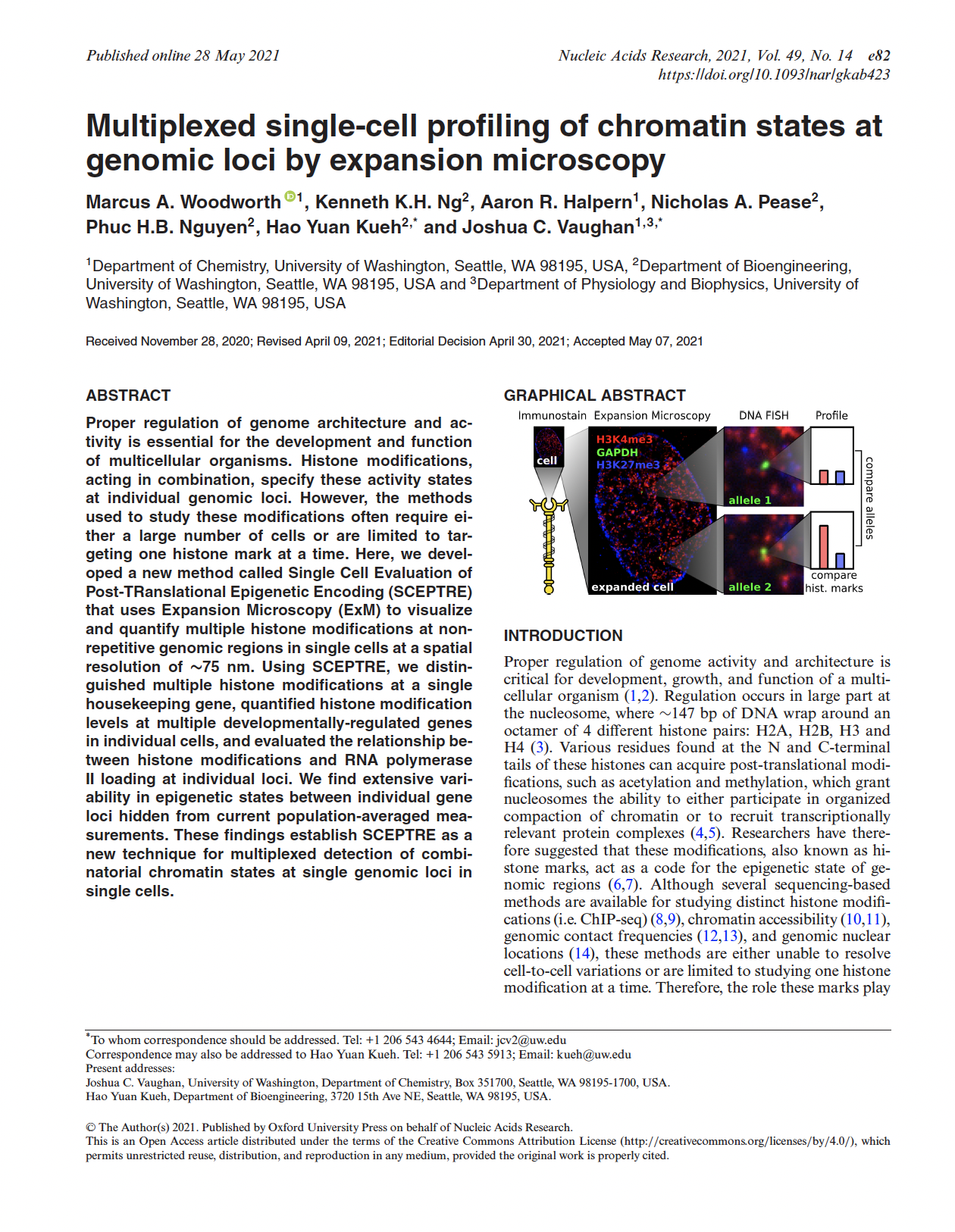
Multiplexed single-cell profiling of chromatin states at genomic loci by expansion microscopy.

Tunable, division-independent control of gene activation timing by a polycomb switch.

Chu J., Pease N.A., Kueh H.Y. (2021). Immunol Rev, 300:134-151. https://doi.org/10.1111/imr.12946

Order by chance: origins and benefits of stochasticity in immune cell fate control.
A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment.
Ng K., Yui M.A., Mehta A., Siu S., Irwin B., Pease S., Hirose S., Elowitz M.B., Rothenberg E.V., Kueh H.Y. (2018). eLife, 7:e37851.
Other publications
Resolving cell cycle speed in one snapshot with a live-cell fluorescent reporter. Eastman A.E., Chen X., Hu X., Hartmann A.A., Pearlman Morales A.M., Yang C., Lu J., Kueh H.Y., Guo S. (2020). Cell Rep, 31(12):107804. https://doi.org/10.1016/j.celrep.2020.107804
Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Longabaugh WJR, Zeng W, Zhang JA, Hosokawa H, Jansen CS, Li L, Romero-Wolf M, Liu P, Kueh HY, Mortazavi A, Rothenberg EV. Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):5800-5807. doi: 10.1073/pnas.1610617114.
A simplified Bcl-2 network model reveals quantitative determinants of cell-to-cell variation in sensitivity to anti-mitotic chemotherapeutics. Kueh H.Y., Zhu Y., Shi J. (2016). Scientific Reports, 6:36585.
Irreversibility of T-Cell Specification: Insights from Computational Modelling of a Minimal Network Architecture. Manesso E., Kueh H.Y., Freedman G., Rothenberg E.V., Peterson C. (2016). PLoS One, 11(8):e0161260.
Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Kueh H.Y., Yui M.A., Ng K., Pease S., Siu S., Bernstein I.D., Elowitz M.B., Rothenberg E.V. (2016). Nature Immunology, 17:956-65. [NI News and Views] [Caltech]
Transcriptional Establishment of Cell-Type Identity: Dynamics and Causal Mechanisms of T-Cell Lineage Commitment. Rothenberg E.V., Champhekar A., Damle S., Del Real M.M., Kueh H.Y., Li L., Yui M.A. (2013). Cold Spring Harb Symp Quant Biol. 2013;78:31-41.
Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Kueh H.Y., Champhekar A., Nutt S.L., Elowitz M.B., Rothenberg E.V. (2013). Science, 341(6146):670-3. [NRI News and Views]
A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Li L., Zhang J.A., Dose M., Kueh H.Y., Mosadeghi R., Gounari F., Rothenberg E.V. (2013). Blood, 122(6):902-11.
Maintenance of mitochondrial oxygen homeostasis by cosubstrate compensation. Kueh H.Y., Niethammer P., Mitchison T.J. (2013). Biophys J, 104(6):1338-48.
Computational modeling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. Manesso E., Chickarmane V., Kueh H.Y., Rothenberg E.V., Peterson C. (2013). J R Soc Interface, 10(78):20120774.
Regulatory gene network circuits underlying T cell development from multipotent progenitors. Kueh H.Y., Rothenberg E.V. (2012) Wiley Interdiscip Rev Syst Biol Med, 4(1):79-102
Actin behavior in bulk cytoplasm is cell cycle regulated in early vertebrate embryos. Field C.M., Wühr M., Anderson G.A., Kueh H.Y., Strickland D., Mitchison TJ. (2011). J Cell Sci. 124(Pt 12):2086-95.
Quantitative analysis of actin turnover in Listeria comet tails: evidence for catastrophic filament turnover. Kueh H.Y., Brieher W.M., Mitchison T.J. (2010). Biophys J, 99(7): 2153-62.
Structural plasticity in actin and tubulin polymer dynamics. Kueh H.Y. and Mitchison T.J. (2009). Science, 325(5943):960-3.
Dynamic stabilization of actin filaments. Kueh H.Y., Brieher W.M., Mitchison T.J. (2008). Proceedings of the National Academy of Sciences, 105(43):16531-6.
Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. Kueh H.Y., Mitchison T.J., Brieher W.M. (2008). Journal of Cell Biology, 182(2):341.
Spatial patterning of metabolism by mitochondria, oxygen and energy sinks in a model cytoplasm. Niethammer P., Kueh H.Y., Mitchison T.J. (2008). Current Biology, 22;18(8):586-91.
Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. Galkin V.E., Orlova A., Brieher W.M., Kueh H.Y., Mitchison T.J., Egelman E.H. (2008). Journal of Molecular Biology. 376(3):607-13.
Intrinsic fluctuations, robustness, and tunability in signaling cycles. Levine J., Kueh H.Y., Mirny L (2007). Biophysical Journal, 92(12):4473-81.
Rapid monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin and Aip1. Brieher W.M., Kueh H.Y., Mitchison T.J (2006). Journal of Cell Biology, 175(2):315-24.
Monitoring actin disassembly with time-lapse microscopy. Kueh H.Y. (2006). Journal of Visualized Experiments, 1:66.
Brain architecture and social complexity in modern and ancient birds. Burish M.J., Kueh H.Y., Wang S.S. (2004). Brain Behavior and Evolution, 63(2):107-24.
Mechanisms of noise-resistance in genetic oscillators. Vilar J.M., Kueh H.Y., Barkai N., Leibler S. (2002). Proceedings of the National Academy of Sciences, 99(9):5988-92.