Research overview
The immune system protects the body against a wide array of threats, ranging from viruses to bacteria to cancer. These versatile defense capabilities are upheld by a collection of immune cell types, each of which can sense and mount responses tailored towards distinct types of threats.
Our lab seeks to elucidate how immune cells fate decisions, both to develop from stem cells, and to mount effective responses to challenges. Immune cells utilize circuits of interacting genes and proteins to sense signals and execute cell state transitions. To understand how these signaling and gene regulatory circuits work, we use powerful reporter systems and quantitative long-term imaging to follow circuit activity in single cells. We also use cutting-edge single-cell genomics and genome engineering techniques to define their molecular parts and architecture, and use mathematical modeling to test and predict their functional performance. Through this work, we aim to lay foundations to rationally program immune cells to fight cancer and other life threatening diseases. Here are examples of recent projects:
Control of T cell lineage commitment by a timed epigenetic switch.
To generate functional T cells, blood stem and progenitor cells must first enter the thymus, where they lose the ability to generate other blood cell types and commit to the T cell lineage. We seek to understand how progenitor cells decide to become a T cell versus other fate alternatives, and how they control these lineage decisions to ensure production of T cells and other immune cell types at the proper numbers and proportions.
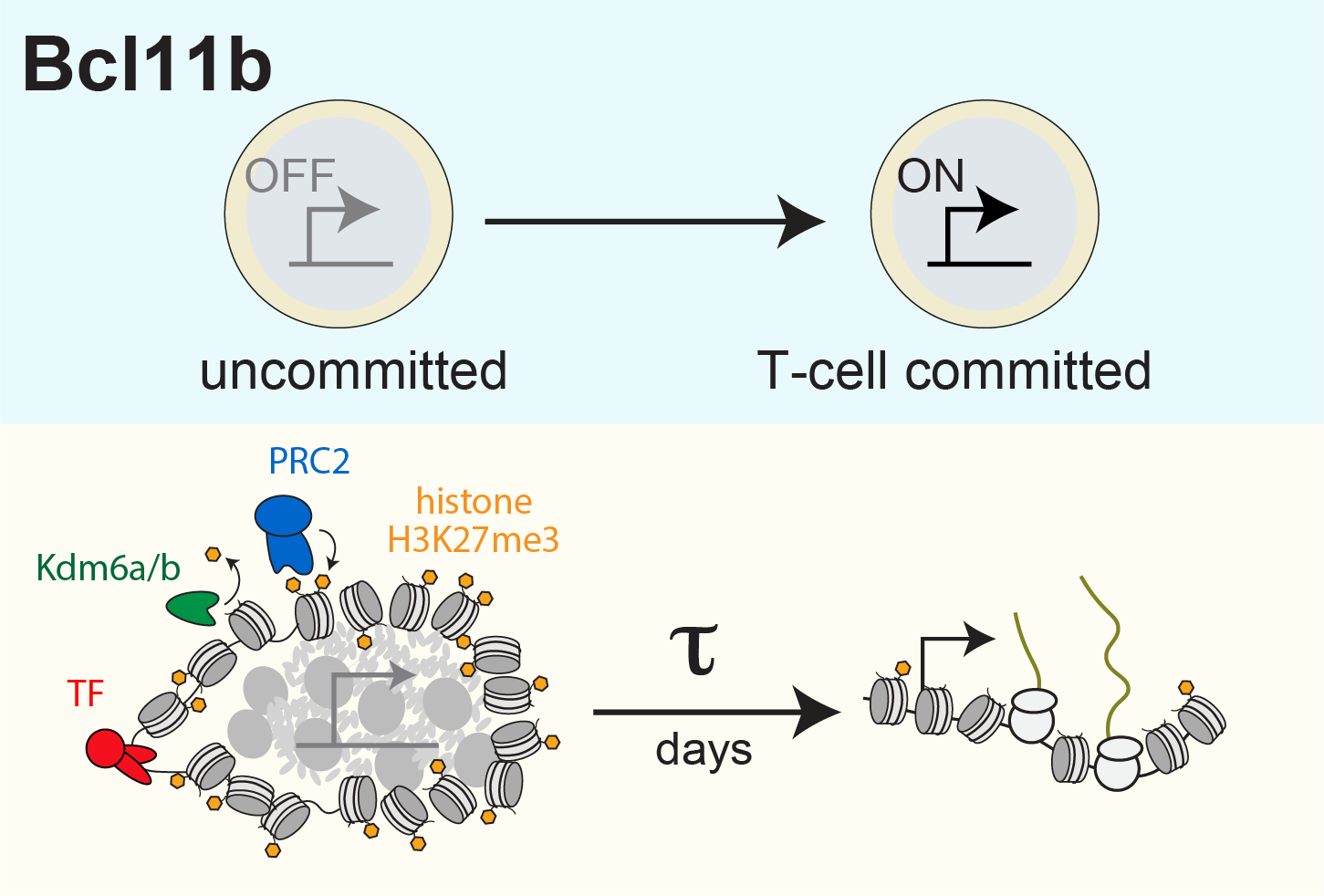
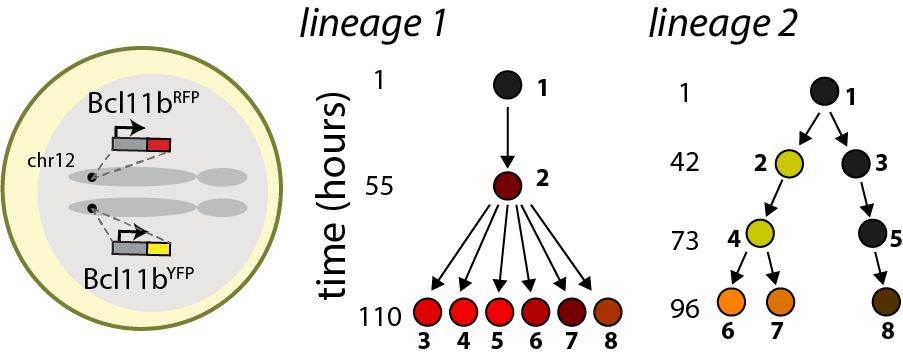
We have uncovered a timed epigenetic switch controlling commitment to the T cell lineage. Progenitors commit to the T cell lineage by turning on the transcription factor Bcl11b, a central regulator of T cell identity (Kueh et al. 2016). We found that the Bcl11b gene locus initially adopts a stable compact chromatin conformation held together by repressive histone methylation, then switches to an extended, demethylated state upon activation and lineage commitment (Pease et al. 2021). Instructive signals in the thymus modulate epigenetic switching; however, by tracking switching dynamics of single Bcl11b loci in living cells, we found that this switch flips with a remarkably long timing delay after signal exposure (Ng et al. 2018), allowing inactive Bcl11b loci to stably maintain a heritable silent state across multiple cell divisions prior to activation. With their flexible operation and their ability to operate over long timescales, such epigenetic switches may broadly modulate cell fate decisions in the immune system and in mammalian development (Nguyen et al. 2021).
We are currently seeking to elucidate the molecular components of this epigenetic switch and the physical basis of its robust, tunable action. We would also like to understand how this switch regulates the numbers of T cells and other immune cells in the body. Finally, we are interested to understand how widely these switches are deployed during immune development and function.
Resolving chromatin modification states at the single cell level.
Covalent modifications of DNA-associated histone proteins are central to the proper expression of lineage-specific genes in response to transcriptional inputs. However, how these chromatin modifications work to control gene expression has been unclear and largely been obscured by current population-averaging methods for chromatin state analysis.
To circumvent these challenges, we have collaborated with the Vaughan lab in UW Chemistry to develop a new imaging-based method for sensitive, multiplexed detection of histone modification states at gene loci in single cells (Woodworth et al. 2021). This method, termed Single-cell Evaluation of Post-Translational Epigenetic states (SCEPTRE), uses expansion microscopy to robustly visualize chromatin states at single gene loci at ~65 nm resolution. Using SCEPTRE, we found substantial cell-to-cell chromatin heterogeneity at both active and silent genes that have been hidden from previous analysis methods.
Using SCEPTRE, we are now seeking to elucidate the extent of chromatin heterogeneity in blood stem and progenitor cells, elucidate the causes of this heterogeneity, and understand the impact of this heterogeneity on lineage choices.
Input discrimination in T cell signaling.
To mount potent yet selective responses against diverse pathogens, T cells sense pathogen-derived peptides with exceptional sensitivity, selectivity and dynamic range: they can detect single peptide copies on target cells while ignoring weaker-binding self-peptides present at much higher levels. At the same time, they discriminate between different peptide numbers over multiple orders of magnitude. We seek to understand how the signaling circuits in T cells have such remarkable sensory capabilities. To do so, we measure signaling activity dynamics in single cells using live reporters, in a system that allows us to precisely alter signaling input strength. We then use these quantitative measurements to be build mathematical models of signaling circuits to explain and predict their performance. This work provides a basis for us to rationally redirect T cell sensing for improved immunotherapies.
T cell effector and memory differentiation.
Upon acute challenge, T cells can differentiate into short-lived effector cells, which have potent anti-pathogen capabilities and memory cells, which can persist to confer long-term protective immunity. The numbers and proportions of effector and memory cells that emerge must be tight controllable to ensure optimal immunity against diverse challenges. We seek to elucidate the pathways and molecular circuitry that underlies effector and memory cell generation. To do so, we are combining cutting edge quantitative imaging and single-cell transcriptomic techniques to track lineage decision making dynamics at the level of clonal cell lineages. This work will allow us to control effector and memory decisions to enhance short and long-term immunity against diverse pathogens.
