SLIMMER
B. Loading DICOM data into RView
1. Run RView
You can run RView either by clicking the RView shortcut, or from the command line.Note for running from the command line:
When running from the command line, use the full path and the binary name, i.e.,
| $ /full/path/to/binary/rviewqt |
2. Using SLIMMER tool to load DICOM data
Select Tools-SLIMMER Motion Correction Tool to load DICOM data into SLIMMER tool.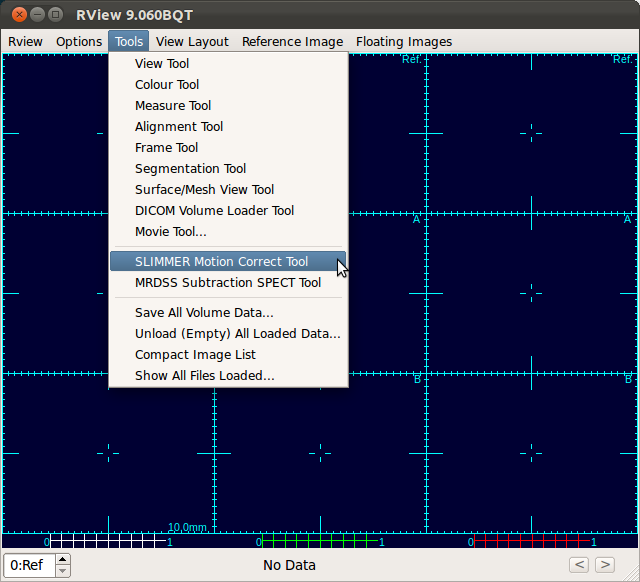
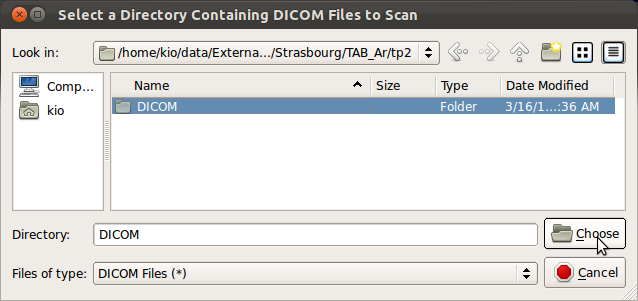
Selecting SLIMMER Motion Correction Tool from the menus will open a directory selection window. Navigate to the top directory that contains desired DICOM files and click Choose button. This wll load all DICOM files within the subdirectories.
3. Selecting stacks to process
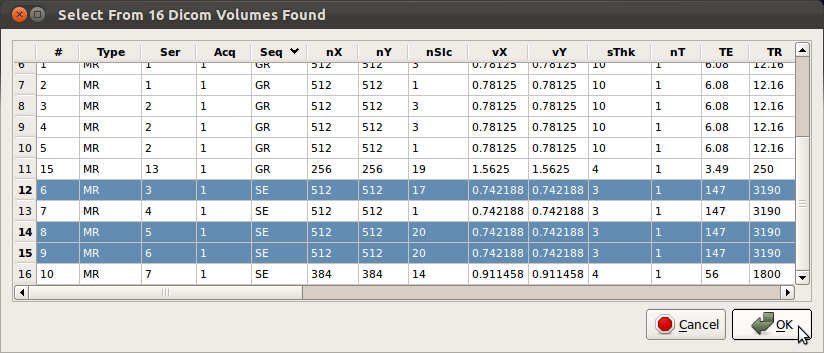
SLIMMER tool is aimed at Fast Spin Echo sequences (SE) such as HASTE or SSFSE, so select only SE sequence stacks.RView will parse all DICOM headers and display available DICOM images grouped by stacks. Seq column displays the sequence code for the stack acquisition, i.e., SE, GR, EP etc. You can find SE stacks by sorting the stacks by the sequence code by clicking Seq column label in the loading window. Check the numbers of slices (nSlc), voxel dimensions (vX, vY, sThk), TR and TE columns to ensure the values are comparable for consistent fusion of the data. Typical values for those variables are shown below.
| Name | Abbrv. | Typical range |
| Number of slices | nSlc | >5 |
| In-plane voxel dimension | vX,vY | <1.5 mm |
| Slice thickness | sThk | <5 mm |
| Echo Time | TE | ~100 ms |
| Repetition Time | TR | >1500 ms |
Once you have selected stacks to load, click OK button to proceed.
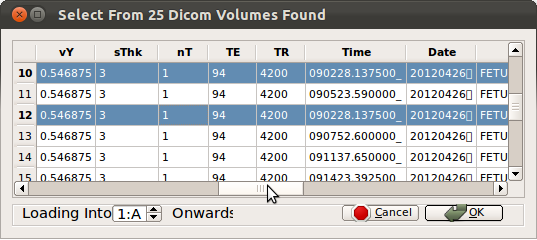
Note: Some systems occasionally make duplicate copies of DICOM data as shown below. Use the horizontal scroll bar and check the acquisition time and date of the stacks to ensure that there exists only one copy at each acquisition time. If multiple copies with identical acquisition time are found, load and inspect them as they could be an original and a post-processed version of the original (for example, distortion or bias corrected version).
Once stacks are loaded, inspect them and remove undesired stacks as explained below. RView will automatically detect and warn if any stacks have identical acquisition time.
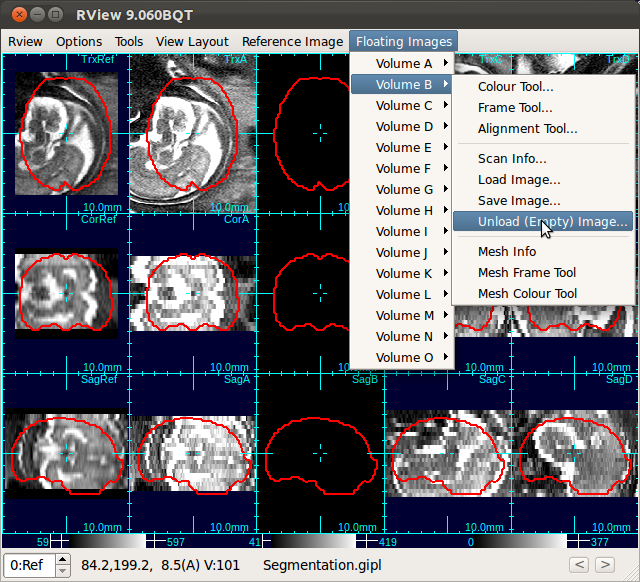
Note: If you want to exclude stacks from the process, you can unload stacks. For example, the stack B in the figure below (the third column) has a substantially different contrast from the rest of the stacks. In order to unload this stack, navigate to Floating Images→Volume B→Unload Image from the menu bar.
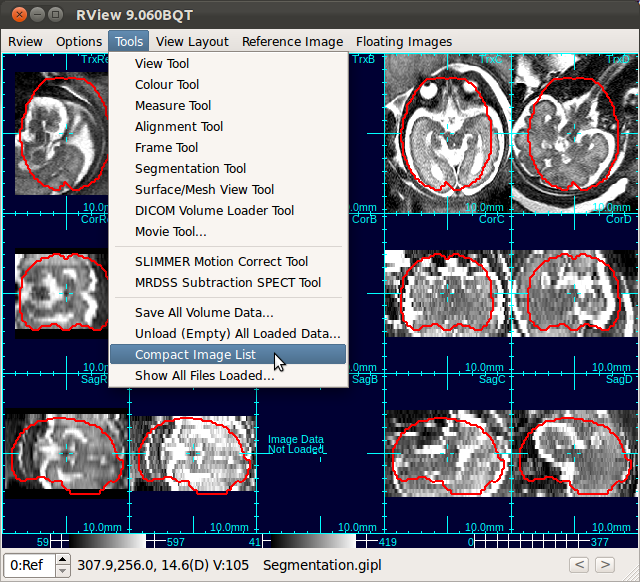
After unloading stacks, select Tools→Compact Image List to the remove blank columns.
«Tutorial Main ‹prev next›
![[UW Pediatrics]](/bicg/images/uwp24.jpg)
