May 13, 2025
Project Highlight: Comparison of Wolf Densities and Diets Across the Prince of Wales Island Complex
By Kylie Baker
Marine Mammals, Genetics, and Scat: Innovative Science Uncovers the Complex Diet and Density of Southeast Alaska’s Wolves
In the rugged forests of Southeast Alaska, a rare species thrives in the shadows, so elusive that even seasoned biologists struggle to find it. The Alexander Archipelago wolf, a small, dark subspecies of gray wolf, is found only in this region and remains one of North America’s most mysterious carnivores. Tracking and studying these coastal wolves has long posed a challenge due to the dense forest and the animal’s evasive behavior.

Yet a groundbreaking collaboration, beginning in 2016, is transforming how we understand their diet, population structure, and resilience in the face of habitat change. This multi-agency effort facilitated through the Pacific Northwest Cooperative Ecosystem Studies Unit (PNW CESU) brings together the Alaska Department of Fish and Game and Oregon State University, with support from U.S. Fish and Wildlife Service-administered Pittman-Robertson funds directed to state agencies. Using cutting-edge genetics, non-invasive sampling, and even scent detection dogs, wildlife researchers are shedding light on a species that has long remained in the dark.
A Conservation Question Spurs Innovation
The project¹ took shape as the U.S. Fish and Wildlife Service weighed whether to list Southeast Alaska’s wolves under the Endangered Species Act. This was due to concerns about habitat loss from logging and the potentially negative effects on Sitka black-tailed deer populations, their primary prey. Dr. Gretchen Roffler, a predator-prey biologist with the Alaska Department of Fish and Game, recognized a major gap in scientific knowledge: how wolf diets vary across this rugged, island-dense landscape. Understanding dietary flexibility was critical for modeling how resilient these wolves might be to ecosystem changes.
Enter Dr. Taal Levi at Oregon State University, whose lab specializes in applying advanced genetic techniques to wildlife ecology. Together with PhD students Charlotte Eriksson and Kayla Fratt, who also brings her expertise as a co-founder of the scent detection dog nonprofit K9 Conservationists, they launched a new approach: using DNA metabarcoding of wolf scat to identify diet composition with far more accuracy and resolution than traditional mechanical sorting.
From Scat to Species: A Genetic Window into Wolf Diets
Fecal DNA metabarcoding involves homogenizing scat samples and applying universal vertebrate primers to extract and amplify DNA from every prey species present. Levi’s lab refined this approach with a custom reference library tailored to the Southeast Alaska ecosystem. Compared to traditional methods that rely on identifying hair and bone fragments, this technique uncovers a much wider array of prey, including soft-bodied and rare species that usually go undetected.
The results were stunning. In analyzing nearly 900 scat samples from across Southeast Alaska, the team identified 55 distinct prey species. While ungulates like deer remained the dominant food source in many areas, in others, wolves surprisingly relied heavily on marine mammals, including sea otters. On Pleasant Island, where wolves recolonized in 2013 and exhausted the local deer population, they made a remarkable dietary switch to sea otters, which had recently rebounded after historic exploitation by the fur trade.

This novel predator-prey relationship was documented in a 2023 publication in Proceedings of the National Academy of Sciences, revealing how wolves adapted to prey scarcity by shifting to marine resources, a finding further corroborated by stable isotope analyses.
Mapping Wolf Populations with Diet and Genetics
To complement dietary analyses, the team developed genomic tools to identify individual wolves from scat using single nucleotide polymorphisms (SNPs), despite the challenge of degraded DNA. PhD student Charlotte Eriksson created a genetic pedigree of the Pleasant Island population, enabling researchers to link individual wolves to specific diets. Intriguingly, the data suggested that on the mainland, female wolves tended to consume smaller prey items—except the breeding female—while Pleasant Island wolves all heavily consumed sea otters.
Building on these discoveries, another Levi Lab PhD student, Ellen Dymit, extended this work to the Alaska Peninsula, where wolves in Katmai National Park were also found to consume large amounts of marine mammals, particularly sea otters. This was a surprising finding, as it showed that the novel predator-prey relationship first documented on Pleasant Island may be generalizable across other parts of the wolves’ coastal range. Meanwhile, in Lake Clark National Park, sea otters are only beginning to recolonize, and wolves have not yet been observed consuming them, though they do prey on other marine mammals. In Kenai Fjords, scat analyses have primarily been identified as coyotes instead of wolves. These findings spurred a new phase of research aimed at understanding how island size and access to marine prey influence wolf density.
Expanding the Scope: Prince of Wales Island and Beyond
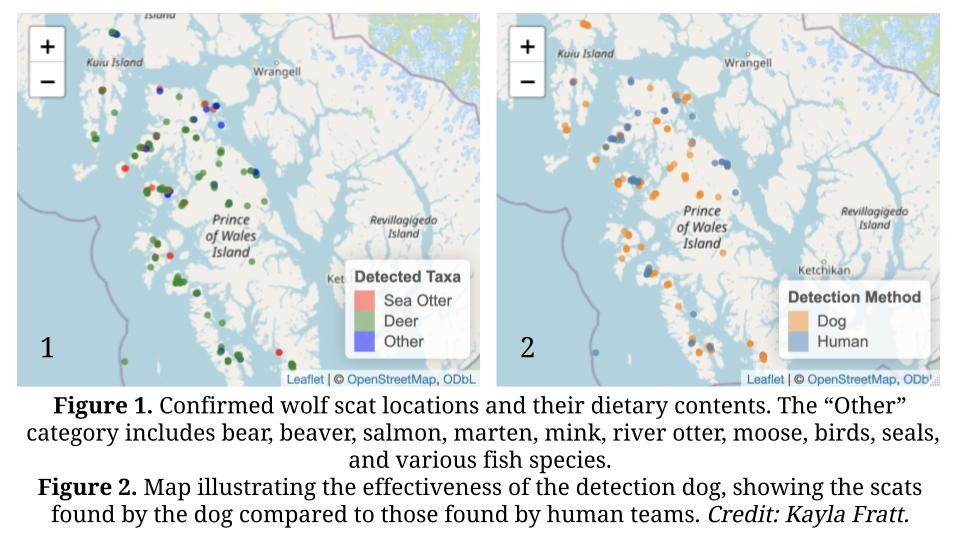
Currently, Kayla Fratt and her scent detection dogs are working alongside technicians from Oregon State and the Alaska Department of Fish and Game to collect wolf scat across the Prince of Wales Island archipelago. The goal: to determine how marine prey availability on smaller, less accessible islands might allow for higher wolf densities, and to improve population estimates across the management unit.

Since 2013, the state has used hair snares and spatial capture-recapture models to estimate wolf abundance, primarily along the island’s road system. However, local communities have raised concerns that this approach underrepresents wolf populations in roadless areas. This new field effort addresses those concerns by targeting remote locations via boat, expanding coverage to include the entire ecosystem.
Next Steps: Toward Genomic Monitoring and Inclusive Collaboration
The team hopes to build on their SNP-based approach to develop a standardized genomic monitoring protocol for wolves in Southeast Alaska and adjacent regions. A more comprehensive SNP panel could enable population tracking over time, help detect potential inbreeding in isolated populations, and facilitate cross-lab collaboration for future studies. Importantly, the data generated through this project supports both state wildlife management decisions—such as sustainable hunting limits—and federal endangered species assessments.
The collaboration between academic researchers, state biologists, federal agencies, and nonprofit partners has already demonstrated the power of emerging technologies to answer pressing ecological questions. With community engagement, logistical ingenuity, and a growing toolkit of molecular methods, this project is redefining what’s possible for wildlife research in even the most challenging landscapes.
¹This initiative has been supported as a PNW CESU project under several project IDs. These include: CT 160001422, CT 180000933, and P20AC00672.
A special thank you to Dr. Gretchen Roffler, Dr. Taal Levi, Charlotte Eriksson, Kayla Fratt, and Ellen Dymit for their critical contributions to this project.