Mentorship Program Experiences
“My experiences in the Research Computing Club’s mentorship program were both fun and extremely valuable to my growth as a mentor. I worked with two fantastic undergraduate students from a different department than my own, which resulted in an impactful multi-disciplinary project related to my graduate research. Coming together with different backgrounds, knowledge and experience strengthened our work together. While the time devoted to different projects varies for different mentor-mentee pairings, my experience specifically as a mentor included about 1 hour a week of in-person meeting time with an additional 1-5 hours a week spent on my own contributions to the project or in other mentorship roles (e.g. providing feedback). What I appreciated about this program is the flexibility in time commitment that it offers both mentors and mentees. In the end, this project resulted in a poster at the UW Undergraduate Research Symposium and the progress made will remain important to the related work in my own research. I would recommend this program to any graduate students interested in improving their mentorship skills while working on impactful research projects with talented students across campus.”
Caitlyn Wolf, Mentor
The RCC Mentorship program is a great opportunity to have undergraduates engaged in research and for graduate students to develop mentorship skills and gain a valuable partner in their projects. Typically when choosing a project, I’ve taken stock of projects I’m working on and determined which of them could most benefit from the skill set the mentee brings to the table. After pitching 2-3 projects we settle on one, and then have weekly meetings for direction and to address obstacles on the project. In the past, I’ve had students work on a variety of projects ranging from installing and benchmarking genome assemblers to training machine learning methods to predict DNA modifications. It’s an excellent chance for undergraduates to get a taste of the scientific process and develop an intuition on how to tackle new problems as they arise. I generally tell students to expect around 5-10 hours a week of research work, although ultimately research is often self-defined in terms of commitment. The more work someone is willing to put in, the more rewarding and higher the chance of getting a publication. For the graduate student, I would expect there to be between 3-5 hours devoted to mentorship and jointly working on the project. I’ve mentored about 7 students and the best ones have always been from the RCC Mentorship program. It’s a great chance to tackle those research projects and ideas that have been shelved but still merit completion.
Aakash Sur, Mentor
Previous Projects
Mentor: Caitlyn Wolf
PI, Dept.: Lilo Pozzo, Chemical Engineering
Research Area(s): Chemical Engineering, Neutron Scattering, Database Management Systems, Cheminformatics, Machine Learning
Undergraduates: Madison Doerr, Yin Yin Low
Programming Language(s): Python
Framework(s): Gaussian, PostgresSQL
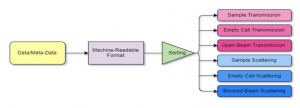
Project Description: Neutron scattering is a powerful, multidisciplinary material characterization tool providing access to molecular-level structure and dynamics. Facilities are generating enormous amounts of data, but it is currently stored disconnected from relevant meta-data. Database management tools would enable a more comprehensive and meaningful arrangement. Critical meta-data, such as sample composition, would then improve analysis of scattering data by use of big data and machine learning (ML) tools. A considerable challenge in developing this database is determining data storage formats that are both descriptive and machine-readable for effective use in ML models. In this work, PostgreSQL and python will be used for development of a neutron scattering database framework and machine learning models. Mentees will gain skills interacting with a database system, managing large amounts of data, developing machine learning models, and using Hyak. Moreover, they will gain exposure to neutron scattering and chemical engineering concepts within the framework of HPC research.
Mentor: Sarah Alamdari
PI, Dept.: Jim Pfaendtner, Chemical Engineering
Research Area(s): Molecular Dynamics Simulations, Ionic Liquids as a solvent for biofuel production, Developing python packages for simulation analysis
Programming Language(s): Python, Bash
Framework(s): Gromacs, Amber, Plumed
Available slots: 2
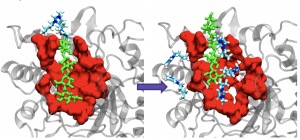
Project Description: Ionic Liquids (ILs) are powerful solvents that can breakdown the highly crystalline structure of non-food plant matter (like poplar cellulose or corn stover), into hydrolyzable sugar molecules that can be fermented and used as biofuels. ILs are an exciting medium as they have been shown to support enzymes in low amounts, a key (and expensive) player in the conversion of cellulose to biofuels. These and many other interesting properties give ILs an economical advantage, that are highly desirable in the biofuel industry. We are interested in creating more resilient and highly productive enzyme/IL systems by understanding their interactions at a molecular level. Two projects are available 1.) Using Molecular Dynamics engines (such as Gromacs and Amber) to perform classical simulations of these systems, recalling important thermodynamic properties. and 2.) Assist in writing a python package for transition path sampling that is indiscriminate to the MD engine (great opp to learn about python and writing packages!)
Mentor: Aakash Sur
PI, Dept.: Peter Myler, Biomedical and Health Informatics
Research Area(s): Genomics, Machine Learning
Undergraduates: Ivan Montero, Frederick Huyan
Programming Language(s): Python
Framework(s): N/A
Research Project: We are a genomics lab, and one of the current projects in the lab is focused on building better genomes for several organisms. That project involves benchmarking various genome assemblers and building data visualizations for large these large datasets. No biology experience is necessary although programming experience is a must. Other projects include using deep learning to train a classifier to learn correct images of genomes, and developing a web-based user-interface for a sequencing database.
PI, Dept.: Mechanical Engineering
Research Area(s): Computational Fluids (oceanography application), algorithm design, postprocessing in parallel
Undergraduates: Yucheng Wang
Programming Language(s): Python, R, C
Framework(s): MatLab
Project Description: Computational fluids is a significant component of the HPC world. My simulations of ocean currents past an underwater mountain will typically output 400GB of data that must be post-processed. On a serial machine, this process typically takes several days. We will discuss data types and RAM limitations for large datasets, and then refactor pre-existing Matlab code into Python, Matlab, Fortran, or C to run on Hyak/Mox. Afterwards, we will have the opportunity to explore the mesmerizing field of flow visualization and use HPC resources to make compelling graphics and/or movies for use in publications.
Mentor: Christopher Nyambura
PI, Dept.: Jim Pfaendtner and Elizabeth Nance, Chemical Engineering
Research Area(s): Molecular Dynamics Simulations
Undergraduates: Jaden Stetler
Programming Language(s): Python
Framework(s): MatLab, Mathematica
Project Description: My project currently involves molecular dynamics simulations of PLGA oligomer chains in various acetone/water solutions. The objective of the mentee would be to setup and run a single PLGA(poly(lactic glycol) acid) chain at specific monomer lengths in water, in acetone and in specific acetone mole fractions. Multiple productions runs will be needed for statistical analysis of macroscopic observables. If these PLGA simulations are easy to setup and run for the mentee, I will also have setup a PEG(polyethylene glycol) chain at specific monomer lengths in water, in acetone and in specific acetone mole fractions. The purpose of these simulations is to ultimately gain molecular level insight into the formulation process of PLGA-PEG nanoparticles via nanoprecipitation.


Recent Comments