 |
|||||||||
|
Research Feature Stories: Other News: |
Teams Generate Models for Protein Folding
A UW research team headed by Dr. David Baker, associate professor of biochemistry and Howard Hughes Medical Institute investigator, was most successful at predicting the three-dimensional structure of a folded protein from a linear sequence of amino acids in the most recent Critical Assessment of Techniques for Protein Structure Prediction (CASP). The biannual CASP competition, which the U.S. Department of Energy initiated in 1994, gave a boost to the field by challenging researchers to produce models without knowing the proteins’ structures.
Baker is a leader in efforts to predict the three-dimensional structures of proteins from their amino acid sequences. He calls protein folding one of the great unsolved mysteries of molecular biology, one that may hold important clues to finding therapies for disease. More than 100 research groups participated in this most recent CASP experiment. They generated possible three-dimensional structures for the 40 candidate proteins selected. The Baker group’s predictions, described as unprecedented in quality, were derived from a computer algorithm the team designed called Rosetta.
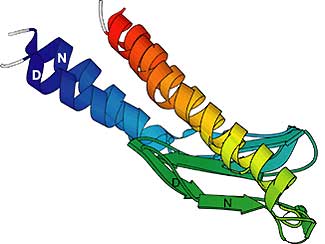 |
This is an example of a structure prediction in the CASP experiment. The predicted structure is shown superimposed on the true structure. The two proteins are colored from blue (start of the sequence) to red (end of the sequence) to facilitate comparison. |
|
| UW AMC Medical Center | UW School of Medicine | Harborview | UW MC | Search UW AMC | UW Home | Contact Us | ©2001-2002, University of Washington Academic Medical Center. All rights reserved. Please honor our copyrights. |
|