Guide to Blood-oxygen-level-dependent i.e., BOLD fMRI, at DISC
Resting state and task-based fMRI is one of the most common sequences used at DISC. Typical scans run from 6 mins to 10 mins. BOLD fMRI is used to assess neuronal activity by measuring change in blood flow and oxygenation. Using this technique brain regions involved in vision, hearing, touch, and emotions can be accessed. Even in the absence of any conscious activity, some regions of the brain are continually "active" and can be mapped using this technique. To enable mapping of regions of interest, DISC provides accessories such as software for stimulus presentation (E-Prime, PsychoPy, Presentation) and technology to support visual, auditory, and tactile cues. We recommend a TE ~ 35 ms, resolution ~ 2-3.5 mm isotropic, MB SENSE = 3~6 or 3-4/R~1-2, latter being preferred based on the latest research findings. Popular parameters for the FMRI sequences at DISC are:
- TR = 1500 ms, TE = 35 ms 3 mm isotropic resolution, no MB SENSE, axial/AC-PC acquisition
- TR = 2400 ms, TE = 15, 30, 45 ms, 3 mm isotropic resolution, no MB SENSE, axial/AC-PC acquisition
- UKBB or Connectome compatible: TR = 700/800 ms, TE = 35 ms, 2 mm isotropic resolution, MB SENSE = 8, axial/AC-PC acquisition
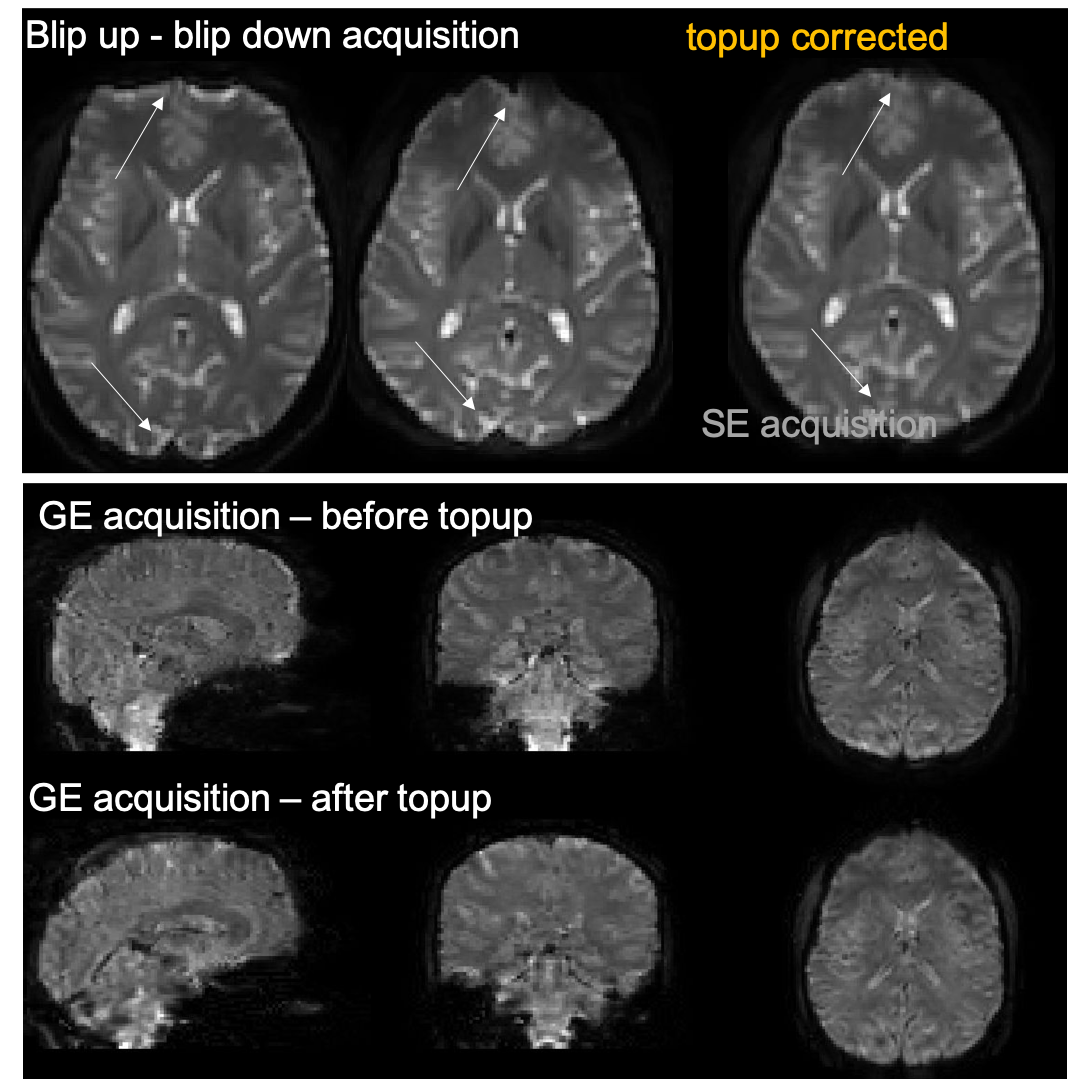
FAQ
- How to calculate echo spacing?
Echo spacing or dwell time = (1000 x water_shift_in_pixels)/(water_fat_shift_in Hz x (epi_factor + 1))/SENSE_factor water shift in pixels is available in the examcard water fat shift in Hz = 3 ppm x 3T x 42.57 MHz/T epi_factor = available in PAR/REC files, DICOM headers and in the exam card SENSE factor = available in the exam card Calculations are in ms.
- In what order are the slices acquired on the Philips scanner? There is no slice timing information in the DICOM header.
Based on the slice scan order, the acquisition can be ascending: 1,2,3,4,5,6,7,8,9,10,11,12; default: 1,3,5,7,9,11,2,4,6,8,10,12; interleaved:1,4,7,10,2,5,8,11,3,6,9,12 (Step by sqrt of total slice no.)
With SMS or MB SENSE = 2, for a
- ascending scan order: 2 parallel acquisitions of (1,7) (2,8) (3,9) (4,10) (5,11) (6,12)
- default scan order: 2 parallel acquisitions of (1,7) (3,9) (5,11) (2,8) (4,10) (6,12)
- interleaved scan order: 2 parallel acquisitions of (1,7) (4,10) (2,8) (5,11) (3,9) (6,12)
With SMS or MB SENSE = 3, for a
- ascending scan order: 3 parallel acquisitions of (1,5,9) (2,6,10) (3,7,11) (4,8,12)
- default scan order: 3 parallel acquisitions of (1,5,9) (3,7,11) (2,6,10) (4,8,12)
- interleave scan order: 3 parallel acquisitions of (1,5,9) (4,8,12) (2,6,10) (3,7,11)
Another example for 30 slices, with MB SENSE = 3, the acquisition for a
default scan order: 3 parallel acquisitions of (1,11,21) (3,13,23) (5,15,25) (7,17,27) (9,19,29) (2,12,22) (3,13,23) (4,14,24) (5,15,25) (6,16,26) (8,18,28) (10,20,30)
In general if no. of slices/MB SENSE is even, that configuration is discouraged due to higher slice leakage artifacts.
- My DICOM data is not sorted by sequences
The Philips DICOM export provides all data in one folder with filenames like IM_*, XX_*, PS_*. To separate the data into separate folders based on sequence, you can use DICOMsort.
For other technical support and learn more about conducting research at DISC, please email DISC Support.
Phone: (206)-685-1604