For the first time, UW ADRC researchers have validated the results of a new systems biology approach to discovering a multi-gene network associated with Alzheimer's disease. They brought computers, worms, and brain tissue to the task.
Genome-wide association studies (GWAS) have so far identified about 20 genetic variants that seem to raise the risk of Alzheimer's disease dementia after age 65. But how do researchers get beyond that first step of gene discovery to an understanding of how a gene variant contributes to disease risk, and then use these variants as potential targets for therapeutics?
In a departure from the one-variant-at-a-time approach, Dr. Shubhabrata Mukherjee, statistical geneticist and Research Assistant Professor, UW Department of Medicine, used a systems biology computational approach to identify— not individual genes—but a network of genes involved in Alzheimer’s risk. A network is a group of genes whose protein products interact within a biological pathway. Mukherjee and his colleagues at the UW Alzheimer's Disease Research Center (ADRC) recently published the experiment's results in Alzheimer's & Dementia.
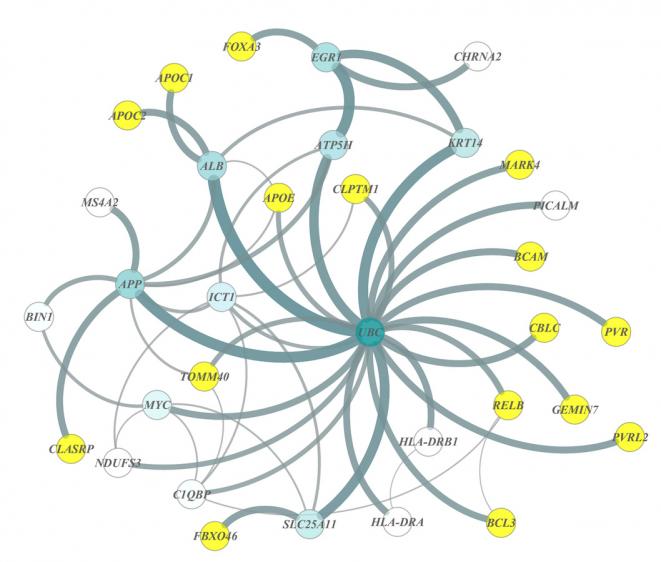
Mukherjee started out by curating the results from the biggest gene variant-wise study of Alzheimer’s disease, a resource compiled by the International Genomics Alzheimer's Project Consortium. The database contains genetic information from tens of thousands of people with and without late-onset Alzheimer's disease. Then, he brought biology into the picture, accessing publically available databases of known protein-protein interactions in humans. His analysis integrated these two sources and produced a chart of the genes overrepresented in Alzheimer’s disease patients and with known connections to each other based on their protein products.
"I was interested to see if we can find a gene that has a lot of other connections," said Mukherjee, "because it could be at the hub of a network of genes that interact with a lot of known genes linked to Alzheimer's disease."
In the newly identified network (see image), the genes serve as nodes in a wide web of connected genes. The hub, or the gene with the highest degree of connections within the network, was UBC, a key gene in the ubiquitin pathway responsible for many biological functions, from DNA repair to cell cycle regulation. It had never been linked to human Alzheimer's risk before, nor had 13 others in this network.
He could have just stopped there. But he wanted validation that this ubiquitin network is truly involved in Alzheimer's disease pathogenesis in a living biological system. However, he couldn't find this answer by staying within his own computational realm. He recalled a conversation with the ADRC's Dr. Matt Kaeberlein, Professor in the UW Department of Pathology, who had offered to help if Mukherjee's group ever unearthed new genes of interest.
Validated at Last
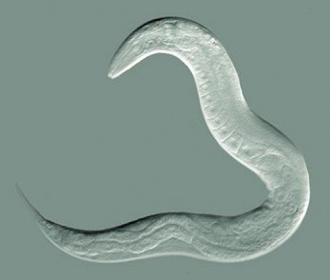
The Kaeberlein Lab uses the Caenorhabditis elegans roundworm as a biological model system for investigating Alzheimer's proteinopathy. These tiny 1-mm worms are genetically manipulated to express the human amyloid beta protein or tau, and they eventually become paralyzed by the toxic effects of these proteins. They can then test the effect of genetic modifiers or drug compounds on the toxicity of these different Alzheimer’s disease pathologies.
When Mukherjee revealed that UBC lies at the center of a large gene network present in Alzheimer's disease patients, Kaeberlein grew excited. His team already had preliminary data showing that the worm version of UBC was a genetic modifier of amyloid toxicity, but they had no evidence that UBC was relevant to the human disease.
"This computational network analysis finding gave me a lot of confidence that the approach of starting from the networks built on human data, and validating them in an invertebrate model, could be a powerful way to identify novel factors involved in fundamental aspects of Alzheimer's disease," said Kaeberlein.
To perform this validation, Dr. Josh Russell, a postdoctoral fellow in the Kaeberlein Lab, tested transgenic worms expressing human amyloid beta under conditions where they produce much less of the worm version of the UBC gene. To do so, he used a technique that interferes with gene expression (RNAi). The worms became paralyzed at an earlier age, suggesting that reduced UBC protein exacerbates the toxicity of amyloid beta. To gain more confidence with these biological results he performed similar experiments to test the four other novel genes identified in Mukherjee's network model. The reduction of each of those genes modified the harmful effects of amyloid beta toxicity.
ADRC collaborators in the Ertekin-Taner lab at Mayo Clinic Jacksonville also analyzed postmortem human brains and found reduced levels of UBC expression in those from people with Alzheimer's disease compared to those from people with other forms of neurodegenerative disease. With these lines of evidence, the research team put forth the hypothesis that UBC is an organizing regulatory molecule in this gene network, which may show decreased activity in people who eventually develop Alzheimer's disease. The hope is that a therapeutic could tweak the upstream or downstream action of the ubiquitin pathway.
"This is just the tip of the iceberg,” said Mukherjee. “Now that it's validated in a biological model of the worm, this UBC gene network is a prime candidate for mechanistic studies to determine how it is influencing Alzheimer's disease-associated pathology. "For example, we're looking forward to the chance to investigate this gene network in an in vitro human model, such as the patient-derived stem cells being created by Drs. Jessica Young and Dirk Keene in the UW Department of Pathology."
In the bigger picture, this two-pronged approach of pairing computational network analysis with biological validation of genetic risk factors is not limited to Alzheimer's disease research. The investigators note that it can also be used to zero in on the genetic networks involved in Parkinson's disease alpha-synuclein pathology and other complex disorders. —Genevieve Wanucha
UW collaborators on this project included Shubhabrata Mukherjee, Josh Russell, Daniel T. Carr, Matt Kaeberlein, Paul Crane, and Thomas J. Montine (former UW ADRC Director, now at Stanford University).