UW CHEMISTRY NMR
Announcments & Equipment Logs
AppNote : GG500 – Effectively Handling 19F baseline
Dear NMR enthusiasts
(Usually the WordPress announcements carry messages about failures and remedies with instruments and carry a tag phrase of ‘Alert’ and ‘Update’. In contrast, this post named ‘AppNote’ carries informtaion/knowledge that you might find useful but you might ignore without much peril ! Hope you find these worth the while)
It is well known that large background signals dominate the baseline of spectra from nuclei such as 19F and 11B. Most of these arise from solid material that is used in the construction of the probe electronics or in the case of 11B, from the NMR tube material itself.
The typical appearnace of these distortinos are in the form of wide humps that appear like ‘hills’ that are tens of ppm broad. Irrespective of the origin of these distortions, the signature of these on the time domain signal i.e. FID is predicatable i.e. large spike like appearance for the first few points of the digitized FID signal. A simple processing technique called ‘backward linear prediction’ can be effectively used to remove baseline distortions caused by these artifacts.
_______________________________________________________________________________________
The good news is that, there is an in-built Topspin automation command that can automagically remove the baseline distortion for you and it uses this very same linear prediction algorithm !
GG500 :
On the GG500 instrument with 19F-optimized iProbe, you can do either of the following to get perfectly flat baselines with 19F
- Automatic : If you start your Flourine experiment by running “fluorine” macro on the command line, at the end of the experiment, simply type : xaup
- the stored Automation Processing script will automatically correct the baseline disortion.
- Manual : If you like setting up the experiment from scratch yourself and process manually, then, after you perform the Fourier Transform, simply run the command : cryoproc1d.
- This command is the same one used in the Automatic method as well.
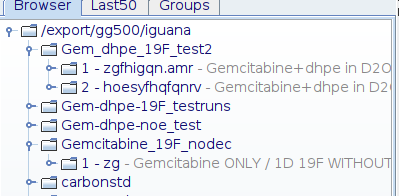
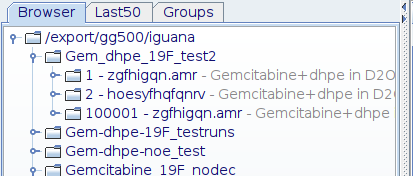
In either of these methods, the script will copy the existing raw data to an unused Exp no. : 100001 (highly unlikely that you would have used this number already). It will then process the data, applying backward linear prediction to give a perfectly flat line.
_______________________________________________________________________________________
Here is some visual guidance, when you attempt this at the instrument :
BEFORE cryoproc1d the file browser looks like this :
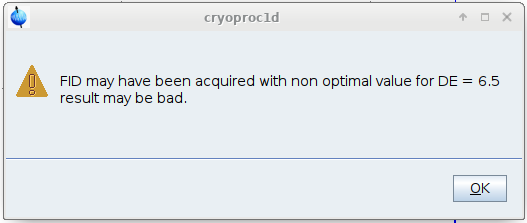
WHEN you run cryoproc1d you will get this warning message, that you can simply acknowledge :
AFTER you run the cryoproc1d, the file browser will show an additional Exp no. where the original FID is saved as a backup.
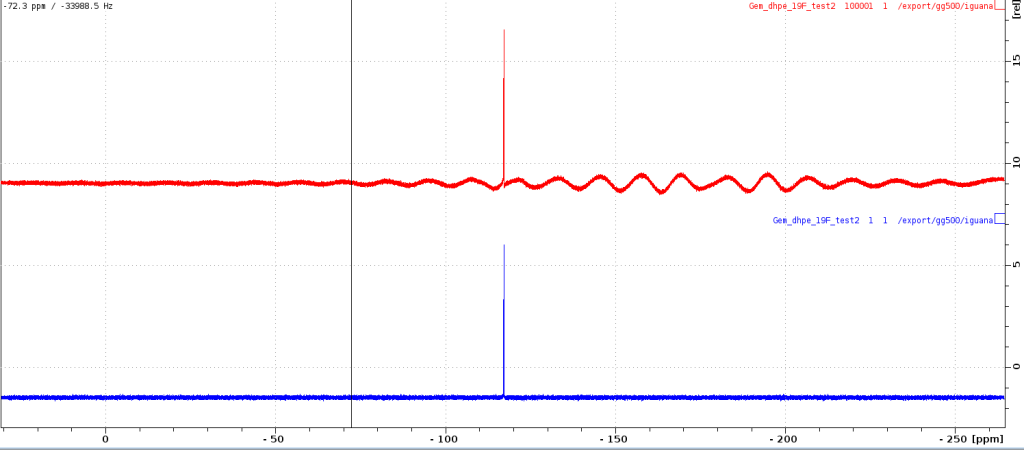
FINAL RESULT of the baseline correction procedure will look something like this. I am adding the spectra that I recorded with my 19F standard on GG500.
- Please note that this is purely a data processing step. So, you can simply re-apply this tool to ALL the raw data that you may have collected on GG500 in the past.
- If you have a version of Topspin 3.x or Topspin 4.x runing on your computer, this command should be in-built.
- The macro ‘fluorine’ and its associated parameter sets have been updated with this automation script so that you can seamelssly use this from now on. When in doubt, simply enter ‘xaup’ on the command line after your ‘efp’ command.
NEO500 :
In short order, we will verify that this automatic baseline cleanup feature is functional in the NEO500 automation setup as well.
Leave a Reply